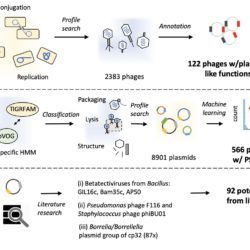
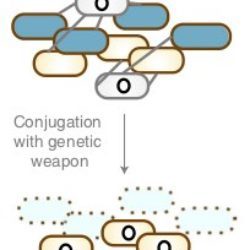
The teams of the department Genomes & Genetics are interested by questions related to gene regulation and evolutionary dynamics, including:
- Replication fidelity.
- Quality control of gene expression.
- Genetic diversity of populations and its impact on adaptation.
- Organisation and evolution of genomes.
This provides solid basis for research programs in synthetic biology, antibiotic resistance, neurodegenerative disorders, and emergent pathogens.
Evolution of virulence, immunity and resistance
The study of how diseases have gradually shaped the human genome over many centuries provides insights on how it may cope with emerging pathogens. By looking into the evolution of infectious microbes and the selective pressures they have exerted on human genes over time, scientists can glean a wealth of useful information to help them understand immune responses. The outcome of the interactions between microbes and hosts is shaped by the genetic diversity of both populations. By improving our understanding of these interactions we obtain vital information on the emergence of novel infectious agents or the increase in resistance to therapies of existing pathogens.
Multidisciplinary approaches
The research conducted in the Department of Genomes and Genetics uses a large panel of multidisciplinary approaches, including molecular genetics, physics, synthetic biology, quantitative biology, genomics, bioinformatics, statistics and proteomics. This includes cutting-edge biophysical methods to produce large-scale analyses of individual or small populations of cells. The department is also the primary scientific affiliation of the Biomics platform that provides high-throughput genomics and metagenomics resources to the campus. These approaches enable us to address all aspects of genome research. Genetics is at the crossroad of many fields of research in biology, and the department is involved in multiple collaborative fundamental and applied research projects. It also hosts in secondary affiliation research labs from almost every other departments in the Pasteur Institute, from virology, microbiology and mycology, to computational biology, structural biology, immunology and global health.
Teaching through research
The department is heavily invested in teaching Institut Pasteur advanced courses. Its members manage the classes on “Multiple roles of RNAs”, “Genome analysis”, “Human Population genomics and genetic epidemiology”, “Mouse genetics”, and “Molecular genetics and epigenetics”. They also teach in many French and foreign universities. The department hosts numerous PhD and Master students from many countries learning how to do research by making it.
Department retreat, Oct 2023 (Pornichet, Loire-Atlantique)