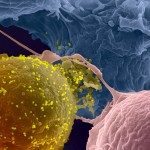
Our work mainly focuses on cellular and molecular aspects
of the replication of HIV and SARS-CoV-2,
and on the interplay between viruses and the host.
We are investigating how pathogenic viruses, mainly HIV, Zika virus and SARS-CoV-2 virus, spread and interact with the host immune system. We are also studying the mechanisms of cell fusion that is either induced by viral infection, or naturally occurs during physiological processes such as placental formation.
Our scientific activities are characterized by three close and complementary axes of research. We are currently studying:
An artistic view of cells and viruses drawings by Fabrice HYBER
When scientists meet artists: http://www.organoide-pasteur.fr
Virus and Immunity Unit
Summer 2023

Summer 2022

March 20, 2020 (lab meeting at the beginning of the lockdown)
June 2019 Lab meeting with Alumni

March 2018
October 2016
October 2014
First Axis
Mechanisms of HIV-1 replication and interaction with the immune system
More than 40 years after its appearance, the HIV/AIDS epidemic still represents a public health threat worldwide. As of 2021, approximately 38 million people are living with HIV/AIDS, with most of them originating from developing countries. Our work focuses on the cellular and molecular aspects of HIV-1 replication, and on the mechanisms of interaction between HIV-infected cells and the immune system. We are currently pursuing three complementary axes of research on this subject:
We are studying viral cell-to-cell spread. Direct cell-to-cell transfer represents a potent and rapid mode of viral propagation, which involves the formation of virological synapses between infected donor cells and uninfected recipients. We are examining the role of cellular and viral proteins which facilitate viral spread. We are deciphering the underlying mechanisms.
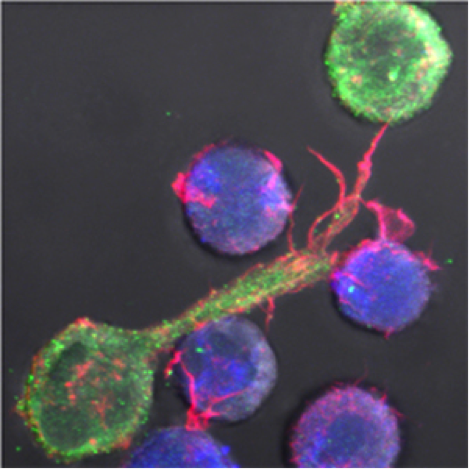
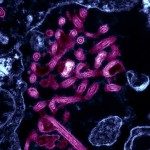
HIV cell-to-cell transfer in lymphocytes (HIV in green, target cell in blue)
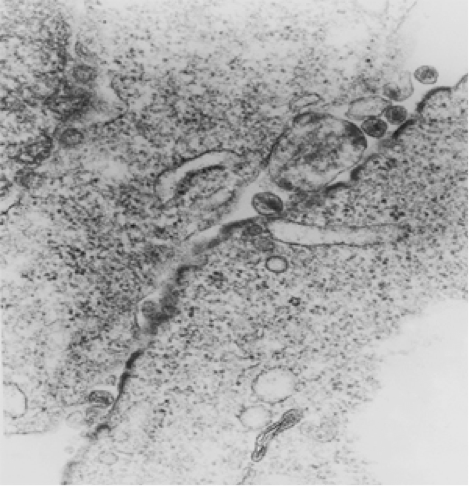
HIV budding at the contact zone between one infected and one target cell
We are also studying the role of the viral protein Nef, which facilitates viral replication by interfering with the antiviral activity of cellular proteins (SERINC proteins), and which also modulates the trafficking of proteins, such as MHC-I that are involved in immune responses.
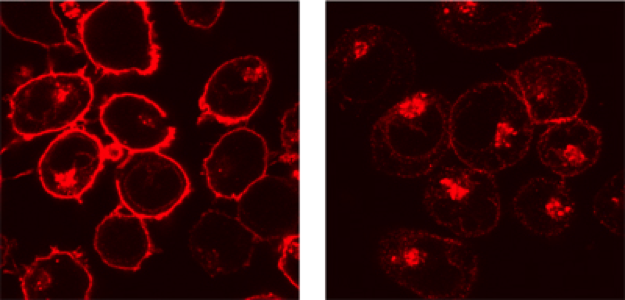
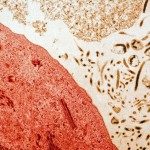
MHC-I intracellular localization in non-infected (right) and Nef-expressing lymphocytes
We are also examining how cellular proteins may inhibit viral spread. These proteins are called restriction factors and are often induced by type-I Interferons. We are studying for instance the role of IFITMs (Interferon induced Transmembrane proteins), which impair viral fusion and then access of incoming virions to the cytoplasm. Our aim is to understand how IFITM proteins inhibit HIV-1 replication and how the virus counteracts this effect.
Movie: Real time imaging of HIV-1 cell-to-cell transmission (from https://www.ncbi.nlm.nih.gov/pubmed/19369333).
Strategies to eliminate the HIV-1 reservoir
In collaboration with the teams of Hugo Mouquet (Institut Pasteur), and Olivier Lambotte (Bicêtre Hospital), we have demonstrated that bNAbs act in complementary ways. Firstly, they neutralize both cell-free virions and cell to cell viral spread. Secondly, the most potent bNAbs bind infected cells and trigger so called effector functions mediate by the Fc domain of the antibody.
We have shown that some bNAbs trigger the destruction of infected cells by Natural Killer (NK) cells through a process termed antibody-dependent cellular cytotoxicity (ADCC). We are currently studying how bNAbs activate the complement pathway at the surface of infected cells.
The complement is a network of proteins initially described for its ability to induce cell toxicity. Due to its ancestral origin, the complement is highly intricated within innate and adaptive immune systems. Complement deposition at the plasma membrane triggers intrinsic signaling pathways impacting cell fate or activation.
Our aims are to study the mechanisms of ADCC and complement deposition by bNAbs and to further assess the ability of bNAbs to clear the HIV-1 reservoir, in cells from patients under successful antiretroviral treatment.
We recently reported that bNAbs also act by tethering budding viral particles at the surface of infected cells, providing an additional antiviral mechanism.
Overall, this project should provide new insights into the activity of antibodies during HIV-1 infection. More generally, it will also help define the mechanisms of antibody-based therapeutic strategies.
Selected publications (see the publication list section, for full reference and link to the articles):
Molinos-Albert LM, Lorin V, Monceaux V, Orr S, Essat A, Dufloo J, Schwartz O, Rouzioux C, Meyer L, Hocqueloux L, Sáez-Cirión A, Mouquet H. Transient viral exposure drives functionally-coordinated humoral immune responses in HIV-1 post-treatment controllers. Nat Commun. 2022 Apr 11;13(1):1944.
Lorin V, Fernández I, Masse-Ranson G, Bouvin-Pley M, Molinos-Albert LM, Planchais C, Hieu T, Péhau-Arnaudet G, Hrebík D, Girelli-Zubani G, Fiquet O, Guivel-Benhassine F, Sanders RW, Walker BD, Schwartz O, Scheid JF, Dimitrov JD, Plevka P, Braibant M, Seaman MS, Bontems F, Di Santo JP, Rey FA, Mouquet H. Epitope convergence of broadly HIV-1 neutralizing IgA and IgG antibody lineages in a viremic controller. J Exp Med. 2022 Mar 7;219(3).
Spencer DA, Goldberg BS, Pandey S, Ordonez T, Dufloo J, Barnette P, Sutton WF, Henderson H, Agnor R, Gao L, Bruel T, Schwartz O, Haigwood NL, Ackerman ME, Hessell AJ. Phagocytosis by an HIV antibody is associated with reduced viremia irrespective of enhanced complement lysis. Nat Commun. 2022 Feb 3;13(1):662.
Dufloo J, Planchais C, Frémont S, Lorin V, Guivel-Benhassine F, Stefic K, Casartelli N, Echard A, Roingeard P, Mouquet H, Schwartz O, Bruel T. Broadly neutralizing anti-HIV-1 antibodies tether viral particles at the surface of infected cells. Nat Commun. 2022 Feb 2;13(1):630.
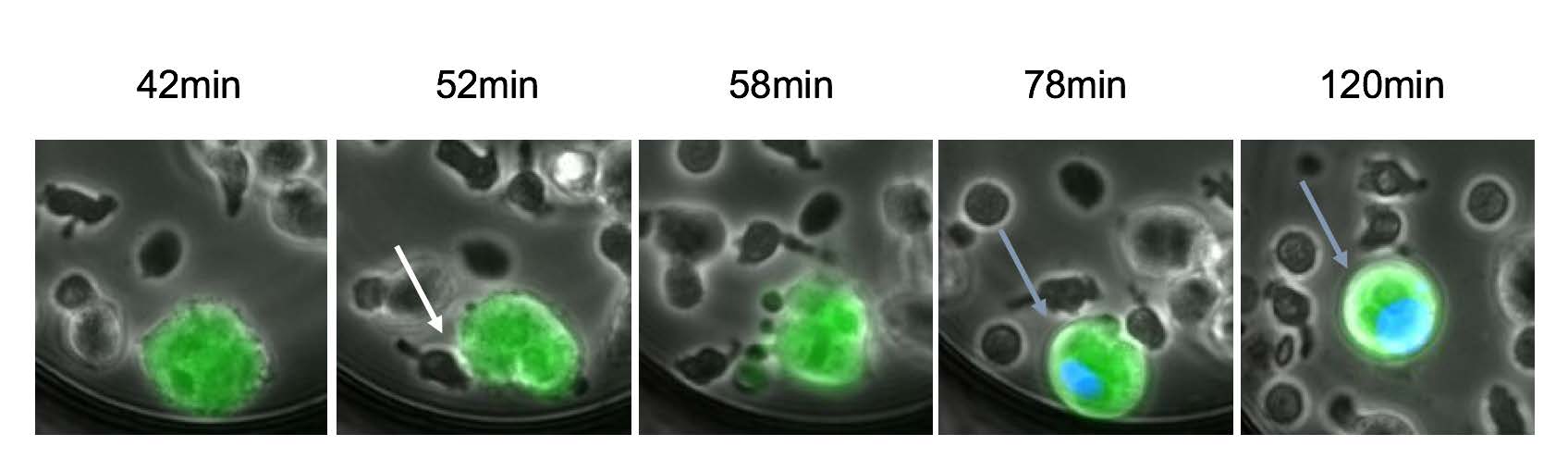
Example of HIV-1-infected cell killing by NK cells: An HIV-1 infected cell (green) and NK (smaller dark cells) are incubated with a bNAb and trapped in a microwells. A picture is taken every 5 minutes. A blue dye reveals the dying cell (Bruel et al. 2016)
Second Axis
The biology and physiopathology of SARS-CoV-2 infection
The COVID-19 pandemic and the continuous emergence of novel variants still pose a significant global public health threat and require an unprecedented research effort in order to provide novel therapeutic and vaccinal strategies. Gaining an understanding of SARS-CoV-2 multiplication and its physiopathology is of the utmost importance.
The innate and adaptative immune response controls viral spread and disease development in most of infected individuals, through mechanisms that are not fully elucidated. Severe cases of COVID-19 are associated with extensive lung damage and the presence of infected multinucleated syncytial pneumocytes. The viral and cellular mechanisms regulating the formation of these syncytia are not well understood. We reported that SARS-CoV-2 infected cells express the Spike protein (S) at their surface and fuse with ACE2-positive neighboring cells (see video below). Expression of S without any other viral proteins triggers syncytia formation. Interferon-induced transmembrane proteins (IFITMs), inhibit S-mediated fusion, with IFITM1 being more active than IFITM2 and IFITM3. On the contrary, the TMPRSS2 serine protease, which is known to enhance infectivity of cell-free virions, processes both S and ACE2 and increases syncytia formation by accelerating the fusion process. TMPRSS2 thwarts the antiviral effect of IFITMs. Our results show that SARS-CoV-2 pathological effects are modulated by cellular proteins that either inhibit or facilitate syncytia formation. We are examining the mechanisms underlying SARS-CoV-2 replication and cell fusion, and the differences that may exist between SARS-CoV-2 variants. We are studying the cytopathic effect of the virus, that is to say the morphological changes in infected cells, in order to understand what happens during the later stages of the viral life cycle. Our aim is to understand how SARS-CoV-2 replicates despite this cytopathic effect, and to further describe the protective innate response of the host. We are using video microscopy, immunofluorescence, electron microscopy and other techniques to address these questions.
Selected publications
Rajah MM, Hubert M, Bishop E, Saunders N, Robinot R, Grzelak L, Planas D, Dufloo J, Gellenoncourt S, Bongers A, Zivaljic M, Planchais C, Guivel-Benhassine F, Porrot F, Mouquet H, Chakrabarti LA, Buchrieser J, Schwartz O. SARS-CoV-2 Alpha, Beta and Delta variants display enhanced Spike-mediated Syncytia Formation. EMBO J. 2021 Oct 2;:e108944.
Rajah MM, Bernier A, Buchrieser J, Schwartz O. The mechanism and consequences of SARS-CoV-2 spike-mediated fusion and syncytia formation. J Mol Biol. 2021 Oct 1;:167280. doi: 10.1016/j.jmb.2021.167280.
Robinot R, Hubert M, de Melo GD, Lazarini F, Bruel T, Smith N, Levallois S, Larrous F, Fernandes J, Gellenoncourt S, Rigaud S, Gorgette O, Thouvenot C, Trébeau C, Mallet A, Duménil G, Gobaa S, Etournay R, Lledo PM, Lecuit M, Bourhy H, Duffy D, Michel V, Schwartz O, Chakrabarti LA. SARS-CoV-2 infection induces the dedifferentiation of multiciliated cells and impairs mucociliary clearance. Nat Commun. 2021 Jul 16;12(1):4354.
Buchrieser J, Dufloo J, Hubert M, Monel B, Planas D, Rajah MM, Planchais C, Porrot F, Guivel-Benhassine F, Van der Werf S, Casartelli N, Mouquet H, Bruel T, Schwartz O. Syncytia formation by SARS-CoV-2 infected cells. EMBO J. 2020 Oct 13:e106267.
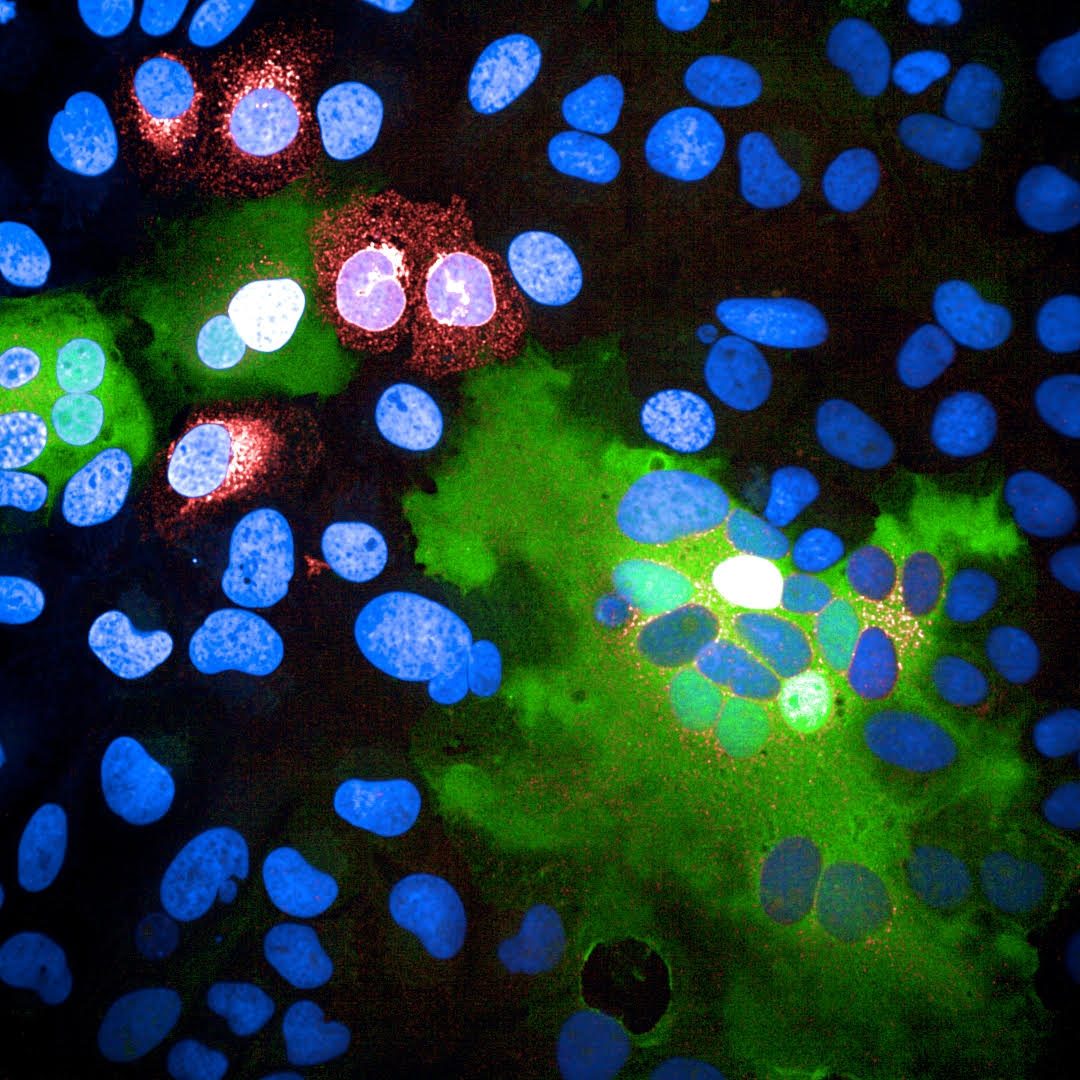
SARS-CoV-2 infected cells fusing with neighboring cells. Red: SARS-CoV-2 Spike protein, Green: Fused cells, Blue: nuclei. (see Buchrieser et al, 2020 for more details)
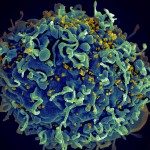
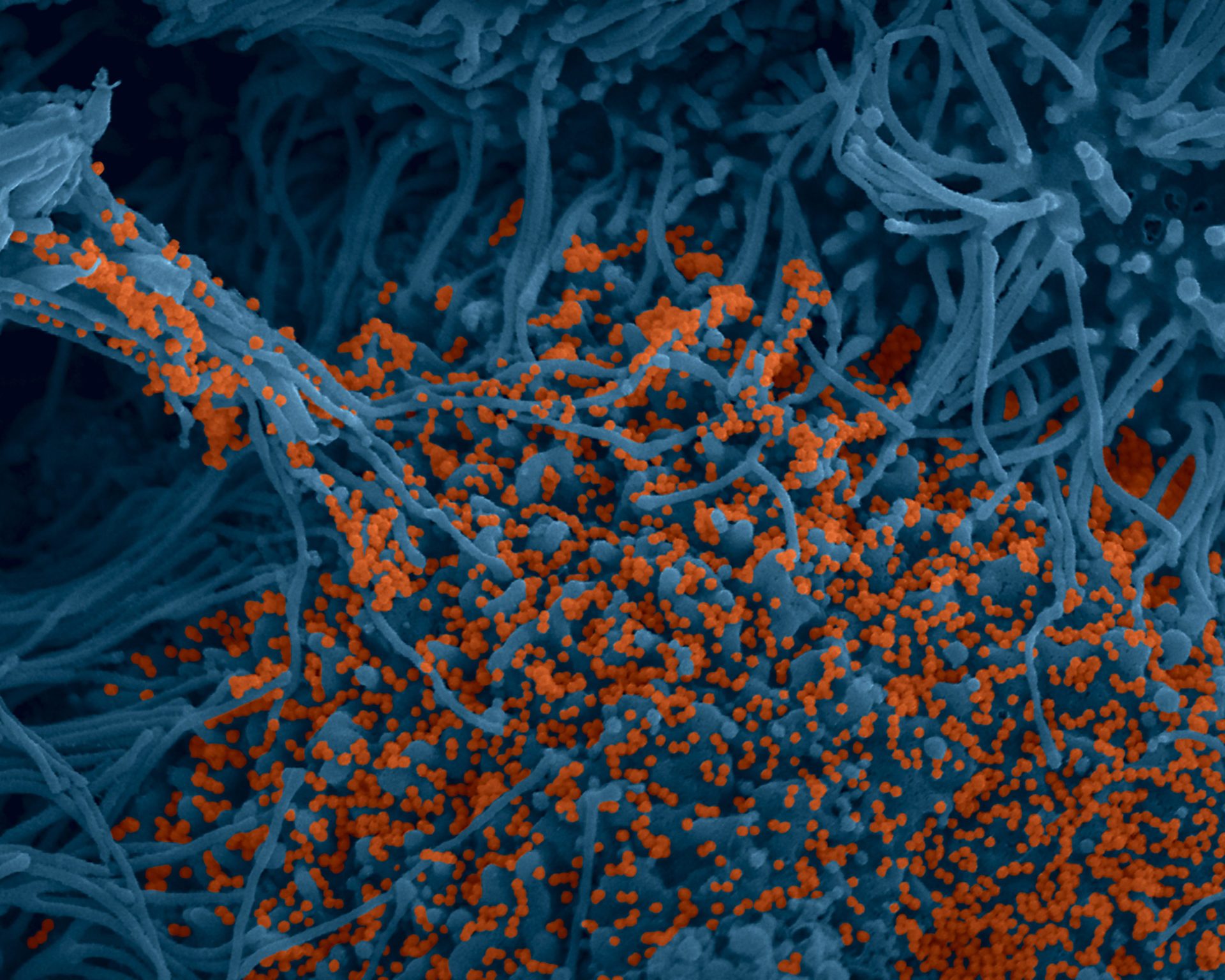
« Human bronchial cells infected by SARS-CoV-2. Scanning electron microscopy. Cells are pseudocolored in blue and virus in orange » (c) Institut Pasteur. Image par Rémy Robinot, Mathieu Hubert, Vincent Michel, Olivier Schwartz & Lisa Chakrabarti, colors by Jean Marc Panaud
Analysis of anti-SARS-CoV-2 antibody function
We have designed cell-based assays to detect, quantify the levels and analyze the activity of anti-SARS-CoV-2 antibodies in infected individuals. The first assay is a flow-cytometry based assay (termed S-Flow) which measures the amounts of anti-Spike protein in serum samples. The second assay (termed S-Fuse), allows the rapid measurement of neutralizing antibodies. We have established a network of collaborations with laboratories in hospitals and institutes to isolate and study emerging variants of concern. We are studying the humoral response to SARS-CoV-2 variants, and the duration of the response, in different categories of infected and vaccinated individuals. We are also assessing the effector function (ADCC, Complement deposition…) of these antibodies.
Selected publications
Planchais C et al. Potent human broadly SARS-CoV-2-neutralizing IgA and IgG antibodies effective against Omicron BA.1 and BA.2. J Exp Med. 2022 Jul 4;219(7):e20220638.
Bruel T et al. Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies. Nature Medicine. 2022 Mar 23;. doi: 10.1038/s41591-022-01792-5.
Simon-Loriere E, Schwartz O. Towards SARS-CoV-2 serotypes?. Nat Rev Microbiol. 2022 Apr;20(4):187-188. doi: 10.1038/s41579-022-00708-x
Planas D et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2021 Dec 23. doi: 10.1038/s41586-021-04389-z.
Planas D et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021 Aug;596(7871):276-280.
Dufloo J et al. Asymptomatic and symptomatic SARS-CoV-2 infections elicit polyfunctional antibodies. Cell Reports Medicine 2021 May 18;2(5):100275.
Planas D et al. 2021. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nature Medicine 2021 Mar 26. doi: 10.1038/s41591-021-01318-5. PMID: 33772244
Third Axis
Cell fusion during placenta formation
High-risk pregnancies occur frequently and may be caused by various factors. It is estimated that 10 to 20% of pregnant women miscarry during their first trimester of pregnancy, and 10% will experience pre-term birth. Slow fetal growth may also arise as a result of maternal infection with certain microbes, parasites or viruses (such as toxoplasmosis or infection with rubella virus, cytomegalovirus, herpes or Zika) or because of genetic or autoimmune diseases. We have identified a new cellular mechanism that alters placental development, potentially causing serious complications during pregnancy. The mechanism is linked with interferon, a molecule produced in response to infection, especially viral infection. The placenta is both a surface for exchange and a barrier between mother and fetus – it delivers nutrients needed for fetal growth, produces hormones and protects the fetus from microbes and the maternal immune system. The external layer of the placenta, the syncytiotrophoblast is composed of cells which fuse together, forming giant cells that are optimized for the placenta’s barrier and exchange functions. If the syncytiotrophoblast fails to form correctly, it can cause placental insufficiency and hinder fetal development.
Interferon is a substance produced by immune cells during infection to combat viruses and other intracellular microbes. High levels of interferon are also observed in autoimmune or inflammatory diseases such as lupus. Exaggerated levels of interferon are responsible for placental abnormality. We demonstrated that interferon induces the production of a family of cellular proteins known as IFITMs (interferon-induced transmembrane proteins), which block cell fusion. Our aim is to understand the mechanism of formation of the placenta under normal or pathological conditions, the mechanism of action of IFN and IFITM, and to study means to prevent placental defects.
Selected publications
Buchrieser J, Schwartz O. Pregnancy complications and Interferon-induced transmembrane proteins (IFITM): balancing antiviral immunity and placental development. C R Biol. 2021 Jul 2;344(2):145-156.
Buchrieser J, Degrelle SA, Couderc T, Nevers Q, Disson O, Manet C, Donahue DA, Porrot F, Hillion KH, Perthame E, Arroyo MV, Souquere S, Ruigrok K, Dupressoir A, Heidmann T, Montagutelli X, Fournier T, Lecuit M, Schwartz O. IFITM proteins inhibit placental syncytiotrophoblast formation and promote fetal demise. Science. 2019 Jul 12;365(6449):176-180.
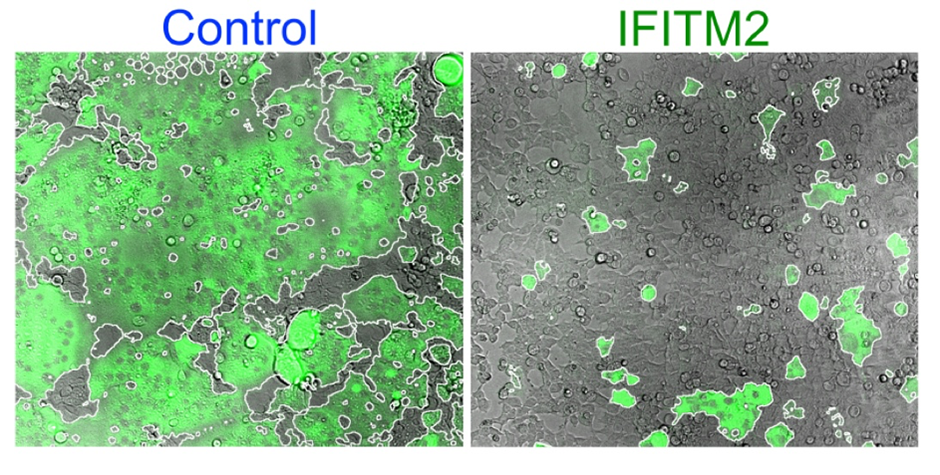
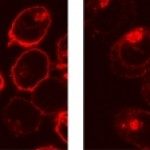
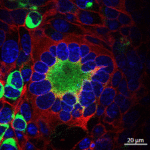
Human cells fusing and forming syncytia (green area) when the Syncytin protein is expressed (left). This process is inhibited by IFITM2 (right). (Buchrieser et al, 2019)
Webmasters : Isma Ziani & Estelle Chaillat
SELECTED COVER PAGES