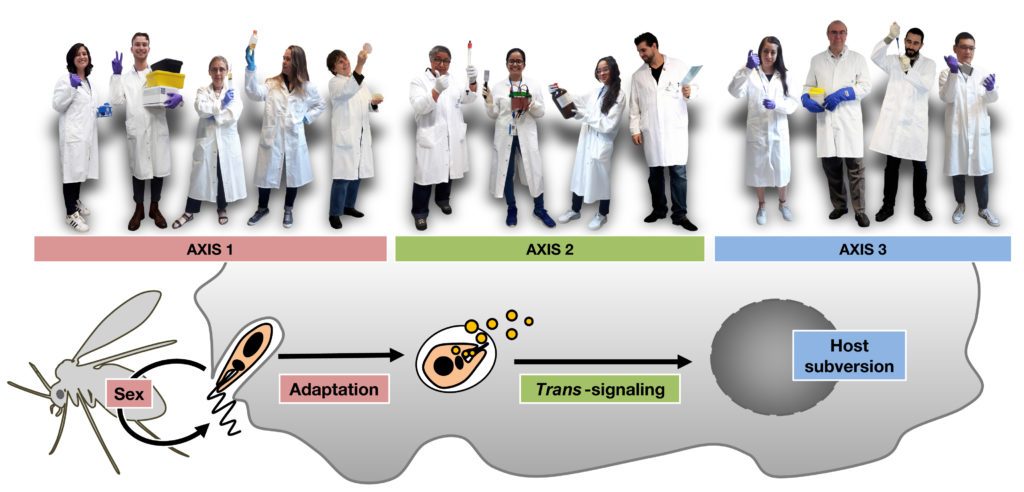
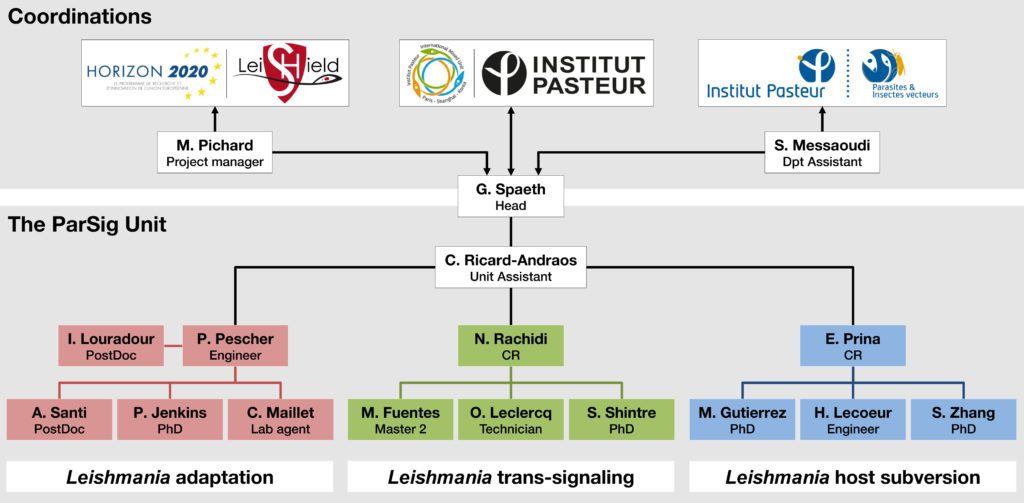
All host/pathogen interactions rely on a constant co-evolutionary battle between genetically highly diverse microbial populations that infect equally diverse human or animal populations, with fitness gains selected both in pathogen and host through complex genotype-genotype interactions. The protozoan parasite Leishmania perfectly well illustrates this genetic cross-talk, with frequent phenotypic shifts (in pathogenicity, drug susceptibility or tissue tropism) selected in Leishmania field isolates through interactions with their heterogeneous vertebrate hosts, which in return react with very diverse clinical responses ranging from asymptomatic infection to fatal immunopathology. Despite the relevance of this highly dynamic host/pathogen relationship for disease outcome and parasite evolution, its underlying molecular mechanisms and physiological consequences remain poorly understood. To address these open questions, our team has initiated over recent years an important thematic transition from its past focus on molecular analysis of parasite signaling to its current and future focus on systems level analysis of Leishmania/macrophage interaction. This new focus animates the projects of three sub-teams that investigate (i) mechanisms of Leishmania evolutionary adaptation and fitness gain at genomic, transcriptomic and translational levels in culture, in animals (sand fly and hamster) and clinical infection (Axis 1), (ii) how Leishmania interferes in trans with macrophage functions through the exosomal release of parasite signaling proteins (Axis 2), and (iii) how Leishmania subverts macrophage immuno-metabolic functions on epigenetic and transcriptional levels to establish permissive conditions for intracellular infection (Axis 3).
Members
Groups

Signalling and host-parasite interactions

Former Members
PostDocs: Suzanne Lamotte (2018 – 2019), Sima Drini (2016 – 2017), Penny Smirlis (2015 – 2017), Mathieu Cayla (2014 – 2015), Mariko Dacher (2012 -2014), Najma Rachidi (2011 – 2015), Wai-Lok Yau (2011 – 2012), Dirk Schmidt-Arras (2007 – 2010), Claire Forestier (2005 – 2008), Miguel Morales (2003 – 2011)
PhD students: Director/co-Director of thesis: Laura Piel (2015 – 2020), Daniel Martel (2015 – 2019), Suzanne Lamotte (2014 – 2018), Sima Drini (2012 – 2016), Mathieu Cayla (2010 – 2014), Mariko Dacher (2008 -2012), Wai-Lok Yau (2009 – 2012)
M2 students: Thibault Rosazza (2018/19), Marie Ajavon (2018), Candide Dossa (2018), Adrien Blisnick (2014), Franck Dumetz (2012), Amel Abdelkarim (2010)
Trainees: Ihcen Kheraci, IP Algers (RIIP* fellow, 3 months 2019); Rafik Garni (RIIP* fellow, 3 months 2019); Arezou Lari, IP Iran (RIIP fellow, 3 months 2019); Farideh Bahari, IP Iran (RIIP* fellow, 3 months 2019); Mariana Boté-Cortes, Fiocruz Brazil (RIIP* visiting fellow, 3 months in 2017); Kyungwa Baek, IP Korea (RIIP fellow, 3 months, 2017); Kossiwa Kokou, IPShanghai (RIIP fellow, 3 months 2016); Evi Gouzelou, Hellenic Pasteur Institute (RIIP trainee, 4 months, 2015); Penny Smirlis, Hellenic Pasteur Institute (EU COST fellow, 1 month, 2015); Joo Hwan No, IP Korea (RIIP fellow, 1 month, 2014); Aymen Bali, (RIIP visiting fellow, 4 months, 2014); Penny Smirlis, Hellenic Pasteur Institute (RIIP trainee, 4 months, 2013); Grace Tewkesbury, Duke University (summer intern Pasteur Foundation, 2 months, 2013); Naouel Eddaikra, IP Alger (RIIP trainee, 1 month, 2013); Monica Gardner, IP Montevideo (RIIP trainee, 2 months, 2012); Sofia Horjales, IP Montevideo (RIIP trainee, 1 month, 2012); Stewart Pine, Harvard University (summer intern, 2 months, 2012); Samin Houshyar, MIT (summer intern, 2 months, 2011); Analia Lima, IP Montevideo (RIIP trainee, 3 months, 2011); Juan-Roman Luque-Ortega, CSIC Madrid (EMBO fellow, 2010); Sylvane Murta, Fiocruz Brazil, (RIIP fellow, 4 months, 2010); Nathalie Joli (summer intern, 1 month, 2010); Najiha Bilal Farooqi (International Fellow, Aga Khan University Medical College, Karachi, Pakistan, 2 months, 2010); Fatma Guerfali, IP Tunis (RIIP trainee, 3 months in 2009); Tahereh Taheri, IP Iran (RIIP trainee, 3 months, 2008), Elahe Davarpanah, IP Iran (RIIP trainee, 3 months in 2022), Ikram Maatallah, IP Morocco (RIIP trainee, 3 months in 2022), Elham Gholami (RIIP trainee, 2 1/2 months, 2023); Yasaman Thaslimi (RIIP trainee, 2 months, 2023, Kahlil Hebbachi ((RIIP trainee, 1 month, 2023); Kamel Benallal ((RIIP trainee, 2 months, 2023)
Jobs
Projects
Transversal Project
Equipments

Cell Imaging System • Life Technologies • EVOS® XL Imaging System
Contact: pascale.pescher@pasteur.fr

Automated Liquid Handling • PerkinElmer • Zephyr Compact Liquid Handling Workstation
Contact: eric.prina@pasteur.fr

Flow cytometer • Beckman Coulter Life Sciences • Cytoflex S (B2Y4) MPL
Contact: najma.rachidi@pasteur.fr

High Performance Electrophoresis • SERVA Electrophoresis • HPE™ FlatTop Tower
Contact: olivier.leclercq@pasteur.fr
Fundings
Publications
Download-
2024Genomic and epidemiological evidence for the emergence of a L. infantum/L. donovani hybrid with unusual epidemiology in northern Italy., mBio 2024 Jun; (): e0099524.
-
2023Intracellular persistence of Leishmania tarentolae in primary canine macrophage cells., Acta Trop 2023 Jul; 243(): 106935.
-
2023Leishmania allelic selection during experimental sand fly infection correlates with mutational signatures of oxidative DNA damage., Proc Natl Acad Sci U S A 2023 Mar; 120(10): e2220828120.
-
2023Amphotericin B resistance correlates with increased fitness in vitro and in vivo in Leishmania (Mundinia) martiniquensis., Front Microbiol 2023 ; 14(): 1156061.
-
2022Trifloxystrobin blocks the growth of Theileria parasites and is a promising drug to treat Buparvaquone resistance., Commun Biol 2022 Nov; 5(1): 1253.
-
2022Retrospective Analysis of Leishmaniasis in Sicily (Italy) from 2013 to 2021: One-Health Impact and Future Control Strategies., Microorganisms 2022 Aug; 10(9): .
-
2022Effects of Structurally Different HDAC Inhibitors against Trypanosoma cruzi, Leishmania, and Schistosoma mansoni., ACS Infect Dis 2022 Jul; 8(7): 1356-1366.
-
2022Going ballistic: Leishmania nuclear subversion of host cell plasticity., Trends Parasitol 2022 Mar; 38(3): 205-216.
-
2022Experimental evolution links post-transcriptional regulation to Leishmania fitness gain., PLoS Pathog 2022 Mar; 18(3): e1010375.
-
2022Released Parasite-Derived Kinases as Novel Targets for Antiparasitic Therapies., Front Cell Infect Microbiol 2022 ; 12(): 825458.
-
+View full list of publications
Trainees
Contact
- Phone: +33 1 40 61 38 58
- Email: gerald.spaeth@pasteur.fr
- Address: 25 – 28 Rue du Docteur Roux, 75015 Paris, France