The Mass Spectrometry for Biology Laboratory is a mixed Institut Pasteur/CNRS Unit (USR 2000) and is headed by Julia Chamot-Rooke (DR1 CNRS). The lab includes the Pasteur proteomics platform and a research group, dedicated to the development of top-down proteomics and structural mass spectrometry. The laboratory is part of the C2RT (Center or Technological Research and Ressources) and of the Structural Biology and Chemistry department. It is IBISA labeled since 2012 and ISO 9001:2015 certified since April 2018.
The platform, headed by M. Matondo, provides service and collaboration in bottom-up proteomics. It is open to external users.
The research group develops four main axes:
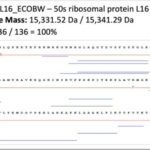
Top-down proteomics is an emerging technology based on the analysis of intact proteins using very high-resolution mass spectrometry. It provides the highest molecular precision for analyzing primary structures and determining post-translational modifications by examining proteins in their intact state, leading to more straightforward and reliable results than the classical bottom-up approach based on protein enzymatic digestion. Top-down proteomics holds great promise in clinical research and in particular in clinical microbiology, a major research axis of the unit.
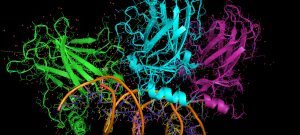
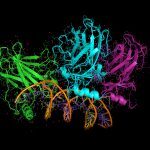
Cross-linking combined to mass spectrometry is a powerful approach to address both the structure and composition of protein complexes. To improve the crucial stage of detection and characterization of cross-linked peptides, we have developed a new generation of cross-linking agents based on click-chemistry.
Native MS allows the structural investigation of protein complexes with the sensitivity, speed, unlimited mass range and accuracy of mass spectrometry. The exact mass of the proteins can be obtained, as well as the stoichiometry of the assembly, its conformation and topological arrangement of the subunits within the complex.
HDX (Hydrogen Deuterium Exchange) MS is based on the different exchange rates of hydrogens in proteins. Exchange rates are a function of solvent accessibility and hydrogen bonding. HDX MS can be used to monitor conformational changes in proteins, characterize protein folding pathways or protein-protein interactions. HDX MS is also widely used for epitope mapping.
For more information on these different topics, please contact Julia Chamot-Rooke