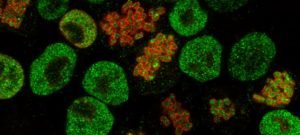
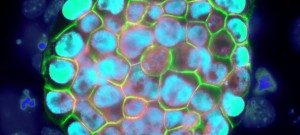
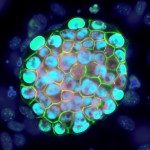
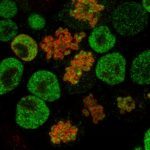
The EPIC unit aims at understanding how transcriptional regulation, chromatin states and signaling pathways functionally interact to bring about and maintain cell identity. Our main model is pluripotency, either in vivo or in the context of self-renewing embryonic stem (ES) cells. Indeed, the establishment, maintenance and execution of pluripotency impose major constraints to gene regulatory processes – understanding the underlying molecular mechanisms is at the heart of our scientific endeavor. Most of our thinking is anchored on the concept of Epigenetics, understood as a way to free gene regulatory processes from the constant need of instructive inputs. In this regard, we want to understand how proliferating cells cope with replication and mitosis such that gene regulatory states are either preserved or changed: what is the role of signaling pathways? How do they connect with Transcription Factors (TFs) and the nucleosomal template? Should a real memory of gene regulation exist, what is its nature? Does it work as a passive storage device or is it rather based on a permanent preservation of regulatory activities?
From this conceptual ground, we have discovered two important regulatory principles in ES cells. First, that repressive histone methylation is strictly dependent on TFs, such as Nanog, Oct4 or Zfp57, and signaling pathways centered on ERK and LIF (Nature Communications 2019, Nucleic Acids Research 2022 and Development 2022). Second, that some TFs such as Esrrb, Nr5a2 and CTCF, display the outstanding capacity to survive replication and mitosis: in contrast to other TFs, they rebind rapidly after the passage of the replication fork and maintain their binding capacity during mitosis, when the chromosomes fully condense. Through their binding, these TFs maintain nucleosome order, whereas at regions losing TF binding the nucleosomes take over and mask TF binding motifs. Hence, Esrrb/Nr5a2 and CTCF confer nucleosome resiliency to replication and mitosis, possibly fostering the reassembly of regulatory complexes in nascent chromatin fibers and in daughter cells (Nature Cell Biology 2016, Genome Research 2019, eLife 2019, Embo Reports 2023 and bioRxiv 2022). Overall, we pioneered the field of mitotic bookmarking factors in ES cells, delivered the first mechanistic basis of their function, and extended their activity to replication, hence producing a new concept, that of “resilient TFs”. Through this work we have not only contributed to a better understanding of how the pluripotency network operates during the cell cycle but also experimentally show the major importance of an under-studied TF, Nr5a2, both in ES cells (Development 2021) and in the embryo, where it executes a chief function to enable the formation of a functional morula (bioRxiv 2023).
Moreover, the group of Michel Cohen-Tannoudji, one of the most recognized French early mouse embryologists, has made key contributions to understand the establishment of the pluripotent epiblast and the primitive endoderm in vivo, including the identification of strict temporal windows of cell responsiveness to inductive cues (Scientific Reports) and intracellular pathways required for lineage stability after specification (Stem Cells).
Together, we are addressing some of the most critical questions in Epigenetics and Developmental Biology: is epigenetic regulation the only mechanism conveying regulatory information to daughter cells? Can the action of transcription factors and long non-coding RNAs be by itself epigenetic? Are histone modifications inherently epigenetic or do they require the constant input of upstream regulators? Are these mechanisms functionally important during normal and pathological processes involving cell proliferation?
This ambitious scientific goal, which requires diverse expertises in molecular, cellular, developmental and computational biology, is supported by both internal (Pasteur, CNRS, Revive labex) and external grants (Consolidator ERC 2018-2023, Ligue Contre le Cancer 2018-2023 and several ANRs).