The Microbial Individuality and Infection (IMI) Unit investigates phenotypic variation in Mycobacterium tuberculosis and the implications for pathogen persistence at the single-cell level, ultimately aiming to improve the control of tuberculosis.
Tuberculosis is a leading global-health threat and a key driver of the antimicrobial resistance crisis. The ability of M. tuberculosis to evade host immunity and to persist despite protracted therapy in a state of alleged quiescence poses a major challenge to the management of this broad-spectrum disease. However, our understanding of pathogen physiology underlying persistence remains incomplete, largely limiting the possibilities of intervention.
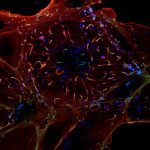
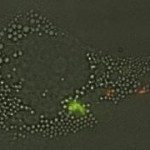
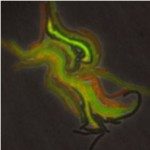
Despite its population clonality, M. tuberculosis exhibits inherent phenotypic variation in fundamental as well as stress-associated cell processes, and develops further cell-to-cell diversity during adverse conditions. This process of diversification is likely to expand its adaptive potential, which we are just beginning to grasp. Our team investigates phenotypic variation in M. tuberculosis using molecular, cellular, omics, and live-cell microscopy approaches. Our central focus is to unravel the mechanisms behind phenotypic diversification, particularly in response to environmental stressors, and to explore how this process contributes to M. tuberculosis persistence. To this aim, we develop cutting-edge time-lapse microfluidic microscopy approaches and combine them with molecular and cell-based functional assays, significantly advancing our understanding of M. tuberculosis adaptative mechanisms. Using low-input RNA-Seq, we identified a subpopulation-specific list of RNA signatures for stress responsiveness that can serve as biomarkers for M. tuberculosis persistence. Drawing from this initial list, we uncovered unexpected relationships between M. tuberculosis phenotypic variation in the expression of certain biomarkers and the ability of specific subpopulations to withstand stressful conditions and to influence the macrophage niche. We also showed that targeting phenotypic variation increases the susceptibility of M. tuberculosis to treatment, paving the way for new therapeutic strategies.
Overall, we believe that a deeper understanding of M. tuberculosis phenotypic diversity will provide critical insights into its survival strategies, and essential clues for the design of pathogen control tools. Our interdisciplinary research approach ultimately aims to develop new therapeutics and diagnostics to improve the management of tuberculosis.
Main IMI’s research objectives:
- Probe the functional role of phenotypic variation in cell-fate determination under host-relevant conditions
One of our primary objectives is to establish the link between phenotypic variation and M. tuberculosis ability to survive under hostile conditions. We dissect the mechanisms underlying M. tuberculosis persistence, with a special focus on DNA and RNA metabolism, molecular chaperones and stress-response pathways, iron homeostasis, cell-wall remodeling, and secreted effectors. Time-lapse microfluidic fluorescence microscopy allows us to capture adaptive responses of single M. tuberculosis cells to stressful conditions. We are also interested in understanding how different M. tuberculosis subpopulations communicate with one another and interact with the host immune system. To this aim, we use a combination of host-cell models, single-cell studies, flow sorting, subpopulation omics, and customized microfluidic devices that simulate the alveolar setting.
- Target phenotypic variation to potentiate therapeutics
We leverage our single-cell findings and understanding of stress-responsive subpopulations, which are more likely to persist, to identify novel therapeutic targets. By hindering specific phenotypic states that confer stress tolerance, we aim to undermine the pathogen and enhance the efficacy of existing anti-tubercular regimens. Corroborating our strategy, we already carried out the first successful single-cell screening for “pheno-tuning” compounds, which decrease phenotypic variation in DNA damage response, increasing M. tuberculosissusceptibility to standard anti-tubercular drugs.
- Clarify the clinical relevance of phenotypic variation via label-free approaches
Our work also extends into the clinical domain, where we assess the presence and relevance of our subpopulation biomarkers of persistence in samples derived from tuberculosis patients. In collaboration with clinicians, we also plan to develop an innovative imaging technology that will increase our understanding of the single-cell biology of M. tuberculosis in the absence of labeling. This approach holds promise for improving early detection, monitoring, and treatment of M. tuberculosis in patients.
In summary, we are implementing a highly interdisciplinary research program, which will enable us to gain a more holistic understanding of M. tuberculosis phenotypic variation and, as a result, to devise new weapons against this dreadful disease.