The UMR3525 studies several aspects of genome biology, with a focus on maintenance and evolution of genetic information. On one hand, most teams work on the organisation, expression, and replication of genomes. On the other hand, most teams study how genomes change through time by mutations, gene acquisition, DNA loss and genetic rearrangements. The key biological models are enterobacteria, Vibrio, and yeast. Recently, the developments in our labs and the integration of the Berthelot team spurred development of projects in humans (and other mammals).
Click to view graph
Connections
Groups
Laboratory
Dynamics of the Genome

Benoit Arcangioli
Laboratory
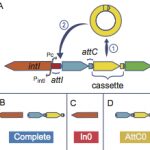
Bacterial Genome Plasticity

Didier Mazel
Laboratory
Microbial Evolutionary Genomics

Eduardo Rocha
Laboratory

Spatial Regulation of Genomes

Romain Koszul
Laboratory
UMR
G5

Comparative Functional Genomics

Camille Berthelot
Laboratory
G5

Molecular diversity of microbes

Aude Bernheim
Laboratory
Synthetic Biology

David Bikard
Laboratory

Sequence Bioinformatics

Rayan Chikhi
Members
Former Members
Former directors of the UMR3525 or URA2171:
- Alain Jacquier (2012-2022)
- Bernard Dujon (2000-2011)
Former principal investigators of the UMR3525 or URA2171:
- Alain Jacquier (Genetics of molecular interactions)
- Bernard Dujon (Molecular genetics of yeast, retired)
- Antoine Danchin (Genetics of bacterial genomes, retired)
- Massimo Vergassola (Physics of the biological systems)
- Frank Kunst (Bacterial genomics, deceased)
- Carmen Buchrieser (Biology of intracellular bacteria)
- Philippe Glaser (Ecology and Evolution of Antibiotics Resistance)
- Roland Brosch (Integrated Mycobacterial Pathogenomics)
Software
Fundings
Publications
Download-
2025Unraveling the Prevalence and Multifaceted Roles of Accessory Peptide Deformylases in Bacterial Adaptation and Resistance., Mol Biol Evol 2025 Nov; 42(12): .
-
2025Beyond RNA modification: a novel role for tRNA modifying enzyme in oxidative stress response and metabolism., Nucleic Acids Res 2025 Nov; 53(22): .
-
2025Structural basis for Lamassu-based antiviral immunity and its evolution from DNA repair machinery., Proc Natl Acad Sci U S A 2025 Nov; 122(47): e2519643122.
-
2025Bacterial natural transformation drives cassette shuffling and simplifies recombination in chromosomal integrons., Nucleic Acids Res 2025 Nov; 53(21): .
-
2025Phages with a broad host range are common across ecosystems., Nat Microbiol 2025 Oct; 10(10): 2537-2549.
-
2025Uridine as a potentiator of aminoglycosides through activation of carbohydrate transporters., Sci Adv 2025 Sep; 11(36): eadw7630.
-
2025Specificity and mechanism of tRNA cleavage by the AriB Toprim nuclease of the PARIS bacterial immune system., Philos Trans R Soc Lond B Biol Sci 2025 Sep; 380(1934): 20240074.
-
2025Antibiotic perturbation of the human gut phageome preserves its individuality and promotes blooms of virulent phages., Cell Rep 2025 Aug; 44(8): 116020.
-
2025RNA Pol II-based regulations of chromosome folding., Cell Genom 2025 Aug; (): 100970.
-
2025Impact of Natural Transformation on the Acquisition of Novel Genes in Bacteria., Mol Biol Evol 2025 Jul; 42(8): .
-
+View full list of publications