 Team
Team
1/ Epigenetics chemical targeting to reprogram cancer cells and human infections
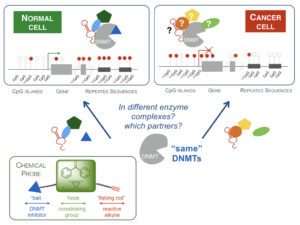
DNA methylation and epigenetic modifications, in general, are reversible and can be modulated by chemical agents. It is in this context we have developed an innovative interdisciplinary approach aiming at the development of new inhibitors of DNA methylation to modulate gene expression. We have validated their use to demethylate hypermethylated gene promoters and reactivate genes in cancer cells. We are investigating the effect of this demethylation on the reprogramming of cancer cells and its consequences. In collaboration we are extending the use of the compounds to other human diseases, as parasite and bacterial infection. The rational is based on the fact that epigenetic changes confer to parasites the phenotypic plasticity, which characterise their progression through the different stages and environments of their life cycle, and bacterial pathogens and viruses alter the epigenetic modifications of the host. 2/ Design and synthesis of novel chemical libraries for epigenetic targets Recent genome-wide data have shown that the key actors of the epigenetic regulation are interlinked and cooperate to induce and maintain aberrant patterns in cancer cells and their microenvironment. An interesting model is thus to address both DNA and histone methylation regulation. We are thus interested in the design of novel inhibitors of HMT (histone methylatransferases). Two are the hurdles identified today: (i) find new chemical scaffolds and (ii) design selective inhibitors when targeting the co-factor pocket, since all share the same co-factor, S-adenosyl-L-methionine (AdoMet). Since the hit rate in screening campaigns against DNMTs and HMTs is particularly low (<0.01%), we are designing an original focused chemical library containing hydrophilic molecules, based on the chemical scaffolds we have developed for DNMTs, that will be tested on a large panel of DNA and histone methyltransferase. 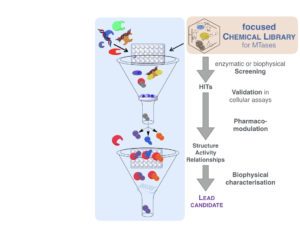
3/ Use of chemical probes to dissect the molecular Epigenetic mechanisms in diseases The chemical tools we design and synthetize will bring an alternative approach to address the biological questions in the understanding of human diseases. For example, an important open question in cancer is what specifically hypermethylates tumor suppressor genes in cancer cells when a global hypomethylation is observed? DNMT inhibitors can be used as chemical probes to address this question. These investigations involve Chemistry and Biology steps: 1) conception and synthesis of chemical probes; 2) trapping experiments for purification of protein as well as for nucleic acid partners; 3) identification of the partners and 4) validation of the identified proteins and nucleic acids as DNMT partners.
Major Collaborations 2010 – 2014 Melanoma field – Dr. G. Favre, ICR, France; Dr L. Lamant, IUC, France; Dr. Luisa Lanfrancone, IEO, Italy; Dr. D. Marzese, JWCI, USA, Prof Carmen Jeronimo, ICBAS, Portugal. 2008 – 2018 Epigenetic field – for chemistry: Dr. D. Guianvarc’h, UPMC, France; Prof. C. Ferroud, CNAM, France; Prof. A. Mai, Università La Sapienza, Italy; Prof. G. Sbardella, Università di Salerno, Italy; Prof. A. Gagnon, UQAM, Canada; Dr. A. Kanamura, U. Oxford, UK; Prof. Gane
san, UEA, UK; – for biology: Prof. A. Jeltsch, Suttgart University, Germany; Dr. J. Tost, CEA, France; Dr L. Mourey, IPBS, France; Prof. P. Jones, Van Andel Institut, USA; Dr M. Frye, U. Cambridge, UK; Prof. B. Huntly, U. Cambridge, UK; Dr A Scherf, I. Pasteur, France. 1999 – 2010 Topoisomerase field – for chemistry: Drs. C. Monneret & D. Dauzonne, I. Curie, France; Drs. A. Garbesi & M. Capobianco, ISOF, Italy; Prof. S. Hecht, Institut, USA; Prof. L. Merlini, Università di Milano, Italy; for biology: Dr. Y. Pommier, NIH, USA; Prof. N. Osheroff, Vanderbilt University, USA; Dr. N. Dekker, Kalvi Institut, TU Delft, The Netherlands
Gupta N, Yakhou L, Albert JR, Azogui A, Ferry L, Kirsh O, Miura F, Battault S, Yamaguchi K, Laisné M, Domrane C, Bonhomme F, Sarkar A, Delagrange M, Ducos B, Cristofari G, Ito T, Greenberg MVC, Defossez PA. A genome-wide screen reveals new regulators of the 2-cell-like cell state. Nat Struct Mol Biol. 2023 Jul 24. doi: 10.1038/s41594-023-01038-z
Andrea Scelfo, Viviana Barra, Nezar Abdennur, George Spracklin, Florence Busato, Catalina Salinas-Luypaert, Elena Bonaiti, Guillaume Velasco, Frédéric Bonhomme, Anna Chipont, Andréa Tijhuis, Diana Spierings, Coralie Guerin, Paola B Arimondo, Claire Francastel, Floris Foijer, Jörg Tost, Leonid Mirny, and Daniele Fachinetti – Tunable DNMT1 degradation reveals DNMT1/DNMT3B synergy in DNA methylation and genome organization – J Cell Biol (2024) 223 (4): e202307026
Yamaguchi K, Chen X, Rodgers B, Miura F, Bashtrykov P, Bonhomme F, Salinas-Luypaert C, Haxholli D, Gutekunst N, Aygenli BÖ, Ferry L, Kirsh O, Laisné M, Scelfo A, Ugur E, Arimondo PB, Leonhardt H, Kanemaki MT, Bartke T, Fachinetti D, Jeltsch A, Ito T, Defossez PA. Non-canonical functions of UHRF1 maintain DNA methylation homeostasis in cancer cells. Nat Commun. 2024 Apr 5;15(1):2960. doi: 10.1038/s41467-024-47314-4. PMID: 38580649; PMCID: PMC10997609