Link to Pubmed [PMID] – 28713344
Link to HAL – pasteur-01555366
Link to DOI – 10.3389/fmicb.2017.01215
Front Microbiol 2017 ; 8(): 1215
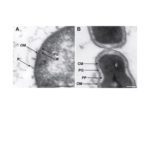
Veillonella parvula is a biofilm-forming commensal found in the lungs, vagina, mouth, and gastro-intestinal tract of humans, yet it may develop into an opportunistic pathogen. Furthermore, the presence of Veillonella has been associated with the development of a healthy immune system in infants. Veillonella belongs to the Negativicutes, a diverse clade of bacteria that represent an evolutionary enigma: they phylogenetically belong to Gram-positive (monoderm) Firmicutes yet maintain an outer membrane (OM) with lipopolysaccharide similar to classic Gram-negative (diderm) bacteria. The OMs of Negativicutes have unique characteristics including the replacement of Braun’s lipoprotein by OmpM for tethering the OM to the peptidoglycan. Through phylogenomic analysis, we have recently provided bioinformatic annotation of the Negativicutes diderm cell envelope. We showed that it is a unique type of envelope that was present in the ancestor of present-day Firmicutes and lost multiple times independently in this phylum, giving rise to the monoderm architecture; however, little experimental data is presently available for any Negativicutes cell envelope. Here, we performed the first experimental proteomic characterization of the cell envelope of a diderm Firmicute, producing an OM proteome of V. parvula. We initially conducted a thorough bioinformatics analysis of all 1,844 predicted proteins from V. parvula DSM 2008’s genome using 12 different localization prediction programs. These results were complemented by protein extraction with surface exposed (SE) protein tags and by subcellular fractionation, both of which were analyzed by liquid chromatography tandem mass spectrometry. The merging of proteomics and bioinformatics results allowed identification of 78 OM proteins. These include a number of receptors for TonB-dependent transport, the main component of the BAM system for OM protein biogenesis (BamA), the Lpt system component LptD, which is responsible for insertion of LPS into the OM, and several copies of the major OmpM protein. The annotation of V. parvula’s OM proteome markedly extends previous inferences on the nature of the cell envelope of Negativicutes, including the experimental evidence of a BAM/TAM system for OM protein biogenesis and of a complete Lpt system for LPS transport to the OM. It also provides important information on the role of OM components in the lifestyle of Veillonella, such as a possible gene cluster for O-antigen synthesis and a large number of adhesins. Finally, many OM hypothetical proteins were identified, which are priority targets for further characterization.