The main focus of the group is to understand the mechanisms of antigenic variation and DNA repair in African trypanosomes, two processes that are intimately linked and essential for virulence. Trypanosoma brucei is a ubiquitous parasite that causes Human African Trypanosomiasis (HAT) in the human host and Nagana in cattle. The broad range of hosts that T. brucei is able to infect make both the medical burden and its economic impact, in terms of daily adjusted life years and rendering large tracts of land unusable for agriculture, devastating to the regions where infection is endemic. Although current treatments suffer from high toxicity and increasing resistance, without treatment HAT is fatal. Trypanosomes are transmitted from the tsetse fly, the insect host, to the mammalian host where they exist and divide in the bloodstream and tissue fluid.
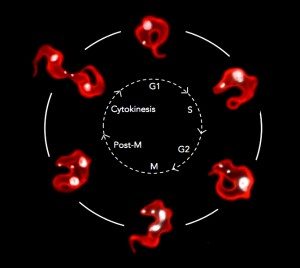
Trypanosomes are covered in approximately 10 million molecules of a single variant surface glycoprotein (VSG), which forms a highly immunogenic surface coat. It is against this coat that the host’s immune response is directed. Antigenic variation is the stochastic switching of the VSG coat, and allows the trypanosome to evade the host’s adaptive immunity. Fundamental to antigenic variation is strict monoallelic expression of a single VSG gene and the concomitant silencing of all others. To date, little is known about how monoallelic expression is controlled in the African trypanosome or how the cells switch VSG`s .
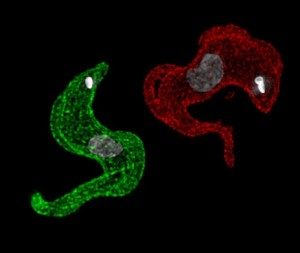
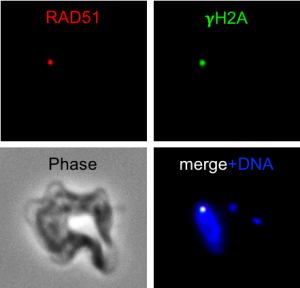
Trypanosomes have evolved to usurp DNA recombination as an essential component of VSG switching. VSG gene conversion events that replace the expressed VSG with one from the VSG archive dominate in T. brucei. These events are dependent on homologous recombination, so in order to better understand antigenic variation we need to better understand DNA recombination and repair. Using established genetic tools we can track DNA repair in T. brucei using DNA damage response markers such as RAD51 and phosphorylated histone H2A. Although homologous recombination appears to be conserved in T. brucei, some components show significant sequence diversity, suggesting functional diversity, which may evolved from the dependence on recombination pathways for antigenic variation.
Using RNA Interference Target screens coupled with NGS (RIT-seq), molecular, biochemical and cell-biological approaches, in high-throughput mode where appropriate, we aim to tackle these longstanding questions that were intractable using prior technologies.