About
Autism spectrum disorders (ASD) are a heterogeneous group of pervasive neurodevelopmental disorders affecting approximately 1% of the population. The diagnosis of ASD is based on impairments in reciprocal social communication and stereotyped behaviors. Behind the apparently unifying definition of ASD lies an extreme degree of clinical and genetic heterogeneity. Indeed, the core symptoms of autism rarely emerge in isolation, but usually coexist with other psychiatric and medical conditions including intellectual disability (ID), epilepsy, motor control difficulties, attention-deficit hyperactivity disorder (ADHD), tics, anxiety, sleep disorders, or gastrointestinal problems. ASD affects more males than females; this is especially true among individuals with a normal intelligence quotient (>5:1) but less so in populations with ID (2:1), and the ratio becomes more balanced if individuals present with dysmorphic features. Individuals with ASD also exhibit alterations in sensory processing, including difficulties in the integration of information across different sensory modalities.
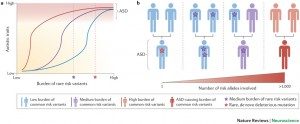
Twin and family studies have conclusively described ASD as the most “genetic” of neuropsychiatric disorders, with concordance rates of 82-92% in monozygotic twins versus 1-10% in dizygotic twins; sibling recurrence risk is 6%. It is now understood that autism symptoms can be caused either by gene mutations or by chromosomal aberrations. In the last years, various independent studies and large-scale international efforts have identified rare variants and copy number variants (CNVs) associated with ASD and suggested a set of mechanisms that could underlie the ASD phenotype. Using current genetic approaches such as single nucleotide polymorphisms (SNPS) arrays, whole exome or genome sequencing, in approximately 10-25% of the cases, a genetic cause can be identified.
At least three main biological pathways have been identified: chromatin remodeling, protein synthesis, synapse formation/function function. Mutations in the genes associated with these pathways represent risk factors for abnormal neuronal connectivity in the brain.
In 2003 we were the first to report mutations of two X-linked neuroligins, NLGN3 and NLGN4X, in individuals with autism or Asperger syndrome. In 2007, the third gene that we identified within this pathway was SHANK3 located on chromosome 22q13, a region deleted in several individuals with ASD. Finally, the fourth gene within this pathway was NRXN1 on chromosome 2p. Neuroligins are cell adhesion molecules with a crucial role in the formation of functional synapses. They are located at the postsynaptic side of the synapse and bind to neurexins located on the pre-synaptic side of the synapse. SHANK3 is a scaffolding protein of the postsynaptic density, which binds to NLGN, and is known to regulate the structural organization of dendritic spines. Since then, we have investigated new synaptic genes associated with ASD such as SHANK1, SHANK2, DLGAP1, CNTNAP4, CNTN5 and CNTN6.
We are currently exploring the interplay between the common and the rare genetic variants on the susceptibility to ASD using next generation sequencing on isolate population (e.g. the Faroese Islands) and in multiplex families. These results strengthen the role of synaptic gene dysfunction in ASD, but also highlight the presence of putative modifier genes, which agrees with a “multiple hit” model for ASD. Our objective is to integrate the genetic and phenotypic data in order to better stratify patients with ASD.