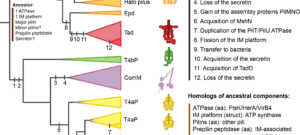
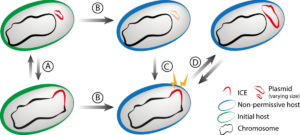
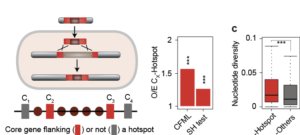
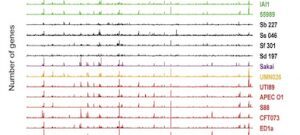
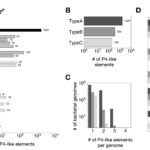
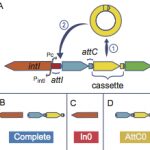
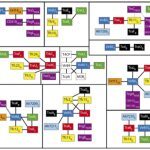
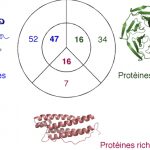
My work focuses on the interplay between the organisation of genomes arising by the chromosomal interactions of cellular processes and the recombination processes generating adaptive genetic variation. Genetic elements tend to be organized in genomes by natural selection in order to optimize this highly complex structure. Yet, they are also constantly faced with genomic variations including mutations, amplifications, rearrangements and horizontal gene transfer. I study the conflict between organisational features and the adaptive plasticity of genomes and how it determines genome evolution and ecological adaptation.
About
Transversal Projects
Projects
Software
Tools
CV
Since 2023 – Head of the UMR3525 CNRS/Institut Pasteur
2019-2023 – Head of the Genomes & Genetics department at Pasteur Institute.
Since 2013 – Head of the unit Microbial Evolutionary Genomics at Pasteur Institute.
Since 2009 – Senior researcher (DR2 then DR1) at the UMR3525 CNRS in Pasteur Institute.
2016-2017 – Member of the COMESP (president SC5)
2015-17 – Associate director for research of the C3BI (Pasteur Institute).
2012-16– President of the CID51 of the National Research Committee (CoNRS).
2010-2015 – Associate director of the GdR BIM.
2008 to 2012– Head junior team Microbial Evolutionary Genomics in URA2171 and UMR3525.
2005– Habilitation in Biology (U Pierre et Marie Curie, Paris).
2000 to 2009– Junior researcher (CR) at the URA2171 CNRS in the Atelier de BioInformatique at UPMC and at Pasteur Institute.
2000– PhD in BioInformatics (U Versailles Saint–Quentin en Yvelines, France).
1997– MSc in Applied Mathematics (Lisbon Technical U, Portugal).
1995– BSc and MSc in Chemical Engineering/Biotechnology (Lisbon Technical U, Portugal).
Publications
Download-
2025Bacterial natural transformation drives cassette shuffling and simplifies recombination in chromosomal integrons., Nucleic Acids Res 2025 Nov; 53(21): .
-
2025Antibiotic perturbation of the human gut phageome preserves its individuality and promotes blooms of virulent phages., Cell Rep 2025 Aug; 44(8): 116020.
-
2025Impact of Natural Transformation on the Acquisition of Novel Genes in Bacteria., Mol Biol Evol 2025 Jul; 42(8): .
-
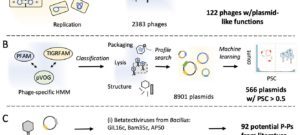
2025Genetics, ecology and evolution of phage satellites., Nat Rev Microbiol 2025 Jul; 23(7): 410-422.
-
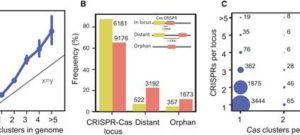
2025Sedentary chromosomal integrons as biobanks of bacterial antiphage defense systems, Science, 388, 6747, (2025)..
-
2025Adaptive genomic plasticity in large-genome, broad-host-range vibrio phages, The International Society of Microbiologial Ecology Journal, In press, pp.wraf063. ⟨10.1093/ismejo/wraf063⟩.
-
2025The Spread of Antibiotic Resistance Is Driven by Plasmids Among the Fastest Evolving and of Broadest Host Range., Mol Biol Evol 2025 Mar; 42(3): .
-
2024Differential stress responsiveness determines intraspecies virulence heterogeneity and host adaptation in Listeria monocytogenes., Nat Microbiol 2024 Dec; 9(12): 3345-3361.
-
2024Expanding the diversity of origin of transfer-containing sequences in mobilizable plasmids, Nature Microbiology, In press, ⟨10.1038/s41564-024-01844-1⟩.
-
2024Intragenomic conflicts with plasmids and chromosomal mobile genetic elements drive the evolution of natural transformation within species., PLoS Biol 2024 Oct; 22(10): e3002814.
-
+View full list of publications