Introduction
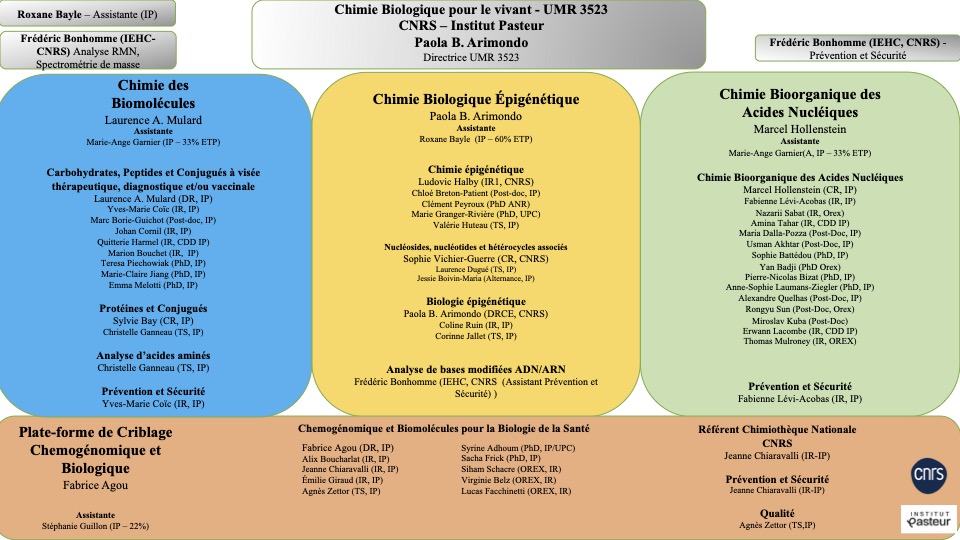
The joint Research Unit UMR3523 CNRS-Institut Pasteur is dedicated to Chemistry for Life (Chem4Life). It is located at the Institut Pasteur in Paris and is part of the Department of Structural Biology and Chemistry. On the CNRS side, Chem4Life is affiliated to the INC as a main institute (section 16) and to the INSB as a secondary institute (sections 21 and 28). Chem4Life is currently composed of 3 research teams dedicated to the chemistry of molecules and biomolecules of biological interest and a technology platform dedicated to assay development and implementation for high-throughput screening (HTS) and high content screening (HCS).
- Chemistry of Biomolecules Unit (Laurence A. Mulard) https://research.pasteur.fr/en/team/chemistry-of-biomolecules/
- Unit of Bioorganic Chemistry of Nucleic Acids (Marcel Hollenstein) https://research.pasteur.fr/en/team/bioorganic-chemistry-of-nucleic-acids/
- Epigenetic Biological Chemistry Unit (Paola B. Arimondo) https://research.pasteur.fr/en/team/chimie-biologique-epigenetique/
- Chemogenomic and Biological Screening Platform (Fabrice Agou) (https://research.pasteur.fr/en/team/fabrice-agou-team/
Each team conducts its research projects independently. Paola B. Arimondo is the administrative director of the UMR 3523. Chem4Life is composed of researchers, engineers and technicians from Institut Pasteur, CNRS and INSERM, and trains students and post-doctoral fellows in both Chemistry and Biology or with a double culture in Chemistry and Biology.
History
Between 2008 and 2016, Chemistry at the Institut Pasteur was represented by two units, the Chemistry of Biomolecules Unit (Laurence A. Mulard, created in 2008) and the Chemistry and Bio-Catalysis Unit (Sylvie Pochet, created in 2010). Recently, Chemistry activities were reinforced to increase the critical mass of research in Chemistry at Institut Pasteur with the arrival of the team of Marcel Hollenstein in 2016, dedicated to the Bioorganic Chemistry of Nucleic Acids and a the Unit Epigenetic Chemical Biology of Paola B. Arimondo in 2018. Sylvie Pochet unit merged into the Epigenetic Chemical Biology one in 2021. To reinforce the drug discovery area, the Chemogenomic and Biological screening platform from the Institut Pasteur (PF-CCB) joint the UMR in January 2022. With these arrivals, the nucleic acid activity has been reinforced, as well as the activity of research into active molecules with therapeutic potential, and Epigenetic Chemistry started with a parallel activity of analysis of modified bases open to all the Ile-de-France teams (EpiK DIMOneHealth project).
Research topics
Chemistry at the interface with Biology has an important role to play at Institut Pasteur, since it opens many possibilities of transdisciplinary, innovative collaborations with biologists. It opens to the development of cutting-edge technological to better understand the biology of human diseases and discover new treatments. The three chemistry teams focus on innovative chemical strategies to elucidate the biological processes that are, in particular, aberrant in diseases; and in parallel they implement new approaches for diagnostic, therapeutic and vaccine applications, together with the core facility dedicated to chemogenomic and biological screening. Chemical probes constitute a valuable toolbox for in detailed studies of the biological processes, whether addressing emerging infectious diseases, AMR, neurodegenerative diseases or cancer. The research projects in Chemistry span from small molecules, bioconjugation methodologies to biomolecules such as nucleobases, nucleosides and nucleic acids, peptides, proteins, carbohydrates and glycoconjugates. The applications go from biosensing of pathogen infections, inflammation and Alzheimer via aptamers, peptides, bioconjugates and chemical probes; sugar derivatives to target the glycan biosynthesis, mimics of nucleosides and nucleobases to interfere with the catalytic activity of enzymes in the replication or translation machinery, to chemical agents that target the Epigenetics, small molecules and peptides to disrupt the DNA-protein and protein-protein interactions and molecular vaccines based on synthetic peptides and glycans as surrogates of bacterial toxins.
Detection and quantification of modified bases in nucleic acids.
Mass spectrometry is a particularly suitable tool to detect and quantify chemical modifications of DNA and RNA. We have developed an experimental protocol based on LC-MS/MS for the relative quantification of modified bases in digested genomic DNA or RNA.
Mass protocols DNA and RNA
500ng-1µg DNA in 20 µL H2O (if necessary we can analyse 200 ng)
Once extracted the DNA with an RNase digestion step or RNA with a DNase digestion step, you need to cut the nucleic acids into nucleosides and purify it. Here is a protocol for that (but many others are available) : use the kit Nucleoside digestion mix of NEB (M0649S) (for DNA after RNase digestion or RNA after DNase digestion).
If you are interested in analysis the global amount of modified bases for your project. The contact for the analysis is Frédéric Bonhomme, EpiCBio lab Institut Pasteur, UMR3523 Chem4Life.
Thanks to numerous collaborations, we now propose to quantify the following modifications
In DNA:
- 5mdC, 5hmdC, 5fdC, 5cadC
- m6dA, 2-aminoadenosine (named Z base), EdA
- BrdU, CldU, EdU
In RNA:
- m5C, m4C, Ac4C, ddhC, Cm
- m6A, m1A, m2A, m6Am, Am
- m1G, m2G, m7G, Gm, Q
- m5U, m3U, Um, Ψ (pseudouridine), D (dihydrouridine) , m1Ψ
Equipment :
Q Exactive™ Hybrid Quadrupole-Orbitrap (Thermo Scientific) mass spectrometer equipped with an electrospray ionisation source, coupled to a Ultimate 3000 RS UHPLC (Thermo Scientific)
Funding :
- Institut Pasteur
- Région Ile-de-France, DIM1HEALTH 2019 project EpiK
Contacts :
Coordinator: Paola B. Arimondo, chem4lifecnrs@pasteur.fr
Partners :
- Institut Pasteur : Chimie Biologique Epigénétique / CNRS UMR 3523
- Institut Pasteur : Biologie des Interactions hôte-Parasite/ CNRS ERL9195/Inserm U1201
- Institut Pasteur : Recherche Dynamique du Génome / CNRS UMR 3525
- Institut Pasteur : Chimie Bioorganique des Acides Nucléiques / CNRS UMR 3523
- Institut Pasteur : Pathogenèse de Helicobacter / CNRS UMR 6047
- Institut Pasteur : Architecture et Dynamique des Macromolécules / CNRS UMR 3528
- Institut Pasteur Dynamique du Génome
- Institut Pasteur : Biologie des ARN de Plasmodium
- Ecole Normal Supérieure : Institut de Biologie / CNRS UMR 8197 / INSERM U1024
- Institut Imagine : Immunité intestinale / INSERM UMR_S1163 / Université de Paris
- INRAE : Epigénétique et Microbiologie Cellulaire
- Université de Paris : Epigénétique et Destin Cellulaire / CNRS UMR7216
- Université de Paris : LCBPT / CNRS UMR 8601
- Institut Pasteur : Plasticité du génome bactérien / CNRS UMR 3525
- Université de Rennes : Institut de Génétique et du Développement de Rennes, CNRS UMR 6290
Core facilities :
Amino Acid Analysis
The Chemistry of Biomolecules Unit offers the qualitative and quantitative determination of the amino acid composition from peptide and protein samples.
Equipment :
VWR Hitachi LA8080 analyzer
Contacts :
Coordinateurs : Sylvie Bay, sylvie.bay@pasteur.fr & Yves-Marie Coïc, yves-marie.coic@pasteur.fr
Réalisations : Christelle Ganneau, christelle.ganneau@pasteur.fr
Chemogenomic and Biological Screening
http://research.pasteur.fr/en/team/fabrice-agou-team/
Teaching activities
- The researchers of the UMR give courses in various universities in France and abroad at different levels, from the bachelor to the doctoral schools.
- In particular, UMR3523 has initiated a new teaching unit dedicated to “Chemical probes and Drug Discovery” which is integrated in the Master Chemistry for Life Sciences, ENS-PSL Sorbonne Université – ChimieParisTech.
- The UMR has also launched the Pasteur Paris University Oxford program which is a PhD exchange programme with the Department of Chemistry of Oxford University in the UK
https://www.pasteur.fr/en/ppu-oxford-program
Jobs
https://research.pasteur.fr/en/search/mulard?pt=+job
https://research.pasteur.fr/en/search/hollenstein?pt=+job
https://research.pasteur.fr/en/search/arimondo?pt=+job
https://research.pasteur.fr/en/search/agou?pt=+job