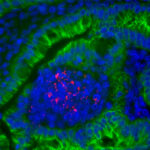
Chromatin modifications, at the level of histones, are fundamental regulators of gene expression in eukaryotes as they control the access of the transcriptional machinery to the targeted promoter regions. Recent studies have found that chromatin modifications induced by bacterial pathogens interfere with the host transcriptional program. However, the mechanisms at play are poorly characterized and the role of these modifications for the host or for the bacterium remain unknown. Research in our team is centered on this new facet of host-pathogen interactions using 2 bacterial models, Listeria monocytogenes and Streptococcus pneumoniae, a pathogen and a natural colonizer respectively. The main goal of our research is to characterize the role of chromatin modifications induced upon bacteria-host interactions and their long term consequences. Our work is at the interface between microbiology, chromatin biology/epigenetics and innate immunity. This multidisciplinary approach will allow the discovery of strategies used by bacteria to reprogram host transcription either during colonization or acute infection, and also provide new insight into fundamental cellular processes such as tolerance to prolonged stimulation and epigenetic memory. In addition, understanding bacteria-induced epigenomic regulation of immune responses could transform our view of immunological memory in vertebrates thereby leading to the development of new antimicrobial agents.
Click to view graph
Connections
About
Members
Former Members
2000
2000
Name
Position
2016
2016
Paul Primard
Medecine student
2017
2018
Kayley Pate
Master student
2015
2018
Jorge Pereira
PhD student
2018
2019
Emma Patey
Master student
2016
2019
Orhan Rasid
Post-doc
2016
2020
Wenyang Dong
PhD student
2019
2021
Matthew Eldridge
Post-doc
2022
2022
Blaise Hebert
Master student
2022
2022
Fanette Seite
Engineer student
2022
2022
Mateusz Checinski
Master student
2019
2022
Tiphaine Camarasa
PhD student
2023
2023
Aymeric Zellner
Licence student
2023
2023
Thomas Harivel
Research Engineer
2016
2023
Marie-Anne LIN
Administrative Assistant
2024
2024
Endre Csaba PAL
Medecine student
2020
2025
Justine Matheau
PhD student
2023
2025
Anna Both
Physician