Biomolecule random walk analysis
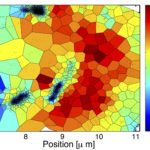
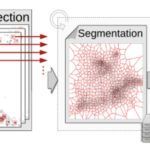
I am in charge of developping the TRamWAy Python library for random walk analysis. The library features multiple tools to build processing chains/pipelines for the spatial or spatio-temporal resolution of physical properties of moving biomolecules such as observed using localization microscopy techniques. These tools include – but are not limited to – tracking biomolecules, segmenting space and time to sample from the identified displacements, and infer local parameters such as diffusivity and effective potential energy or force/drift. This latter inference step is based on a model that derives from the overdamped Langevin equation. In addition, these computation steps can be assembled into a pipeline and TRamWAy can dispatch the computations from a local Jupyter notebook onto a remote high-performance-computing cluster such as Maestro (at Institut Pasteur). I have also developed various analysis applications based on TRamWAy in the context of projects such as Mapping Receptor Dynamics in Synapses. I took part in collaborations that aimed at analyzing the dynamics of GlyB receptors in synapses, of Gag proteins at the membrane of CD4+ lymphocytes, and I still contribute to studies on the formation of Virus-Like Particles, with localization microscopy data for various molecules such as Tetherin, GT46 and CD59.
Modelling of the Drosophila larva behaviour
I have taken an increasingly active part in the main research axis of the G5 Decision and Bayesian Computation group: Deciding in Complex Environments. I first brought technical support to various collaborating groups with the deployment of the behaviour-tagging pipeline solution developed by Jean-Baptiste Masson. I did so at Tomoko Ohyama’s lab at McGill University, Montreal, Canada, and at Marta Zlatic’s lab at the Laboratory of Molecular Biology in Cambridge, UK. I also implemented extensive parts of the data management and data processing automated pipeline at Tihana Jovanic’s lab at NeuroPSI in Gif-sur-Yvette, France, from the experimental setup (acquisition station) to Institut Pasteur, and then back to NeuroPSI. Lately, I am working on building a dictionary of behavioural responses using string metrics on string-like representations of the responses.