Présentation
The single cell transcriptomics, epigenetic and genomics are supported by collaborative effort of the CB UTechS, Biomics and the Hub.
The CB UTechS gathers expertise and high-end instrumentation for single cell phenotyping and sorting, single cell transcriptomics, epigenetic, genomics and single cell data analysis. Biomics proposes up-to-date solutions for Next Generation Sequencing (NGS), with an autonomous access to sequencers for sequencing of single cell libraries. Together, we offer an end-to-end solution from study design, cell enrichment by FACS, single cell isolation and library preparation to NGS and bioinformatic data analysis. The team of dedicated professionals provides expert advice on experimental design, instrument training, data analysis and support to users.
Applications such as single cell transcriptomics, epigenomics, TCR/BCR immune profiling, targeted single cell gene expression (qPCR) are proposed. Development of novel pipelines are available on request.
Technologies
The field of single cell transcriptomics, epigenetic and genomics is the most rapidly evolving of all omics approaches. It consists of various commercial technologies and non-commercial pipelines. The solutions available at the CB UTechS are described below:

10X Chromium. 10x Genomics Chromium technology partitions reactions into microfluidic droplets containing uniquely barcoded beads called GEMs (Gel Bead-In EMulsions). This core technology can be used to partition single cells or nuclei to prepare next generation sequencing libraries in parallel. The 10xChromium is couplet to automated library preparation using Bravo Agilent liquid handling platform. Main features of the 10xChromium are described here. Available applications: Gene Expression 3′ and 5′, Single Cell Immune Profiling, CITE-seq, ATAC-seq, Visium Spatial Transcriptomics.
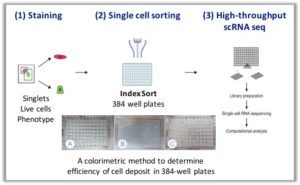
MARS-Seq. The Massively parallel single-cell RNA-sequencing (MARS-Seq) method is based on FACS sorting of single cells into 384-well plates and subsequent automated processing that is done mostly on pooled and labeled material. This methodology uses three levels of barcoding (molecular-, cellular-, and plate-level tags) to facilitate molecule counting with a high degree of multiplexing.
 Customised Microfluidics (technology development). In collaboration with Dynamics of Host Pathogen interaction Unit (headed by J. Enninga), and based on technologies developed by ESPCI we are developing solutions for single eucaryote/procaryote sorting and transcriptomics.
Customised Microfluidics (technology development). In collaboration with Dynamics of Host Pathogen interaction Unit (headed by J. Enninga), and based on technologies developed by ESPCI we are developing solutions for single eucaryote/procaryote sorting and transcriptomics.
 Biomark, Fluidigm. The Biomark can run microfluidic chips (Integrated Fluidic Circuits, IFC) for massively parallel QPCR of up to 96 cells, targeting up to 96 genes. The system utilises low volumes of standard gene expression for genotyping TaqMan or Evergreen systems. Different IFCs format are available to detect 48 genes in 48 cells, or 96 genes in 96 cells.
Biomark, Fluidigm. The Biomark can run microfluidic chips (Integrated Fluidic Circuits, IFC) for massively parallel QPCR of up to 96 cells, targeting up to 96 genes. The system utilises low volumes of standard gene expression for genotyping TaqMan or Evergreen systems. Different IFCs format are available to detect 48 genes in 48 cells, or 96 genes in 96 cells.
 C1, Fluidigm. Microfluidic-based technology that enables automated on-chip single cell separation, reverse transcription and pre-amplification using just one technology and thus reducing the variability caused by multi-platform technical errors. It supports multiple single-cell applications, allowing full-length RNA-seq, targeted gene expression, miRNA analysis, DNA-seq and ATAC-seq.
C1, Fluidigm. Microfluidic-based technology that enables automated on-chip single cell separation, reverse transcription and pre-amplification using just one technology and thus reducing the variability caused by multi-platform technical errors. It supports multiple single-cell applications, allowing full-length RNA-seq, targeted gene expression, miRNA analysis, DNA-seq and ATAC-seq.
Quality control and quantification
 Bioanalyzer uses a micro-capillary based electrophoretic cell that allows rapid and sensitive quantification, quality control and size analysis of DNA and RNA. . TapeStation is an automated electrophoresis solution for the sample quality control of DNA and RNA samples. Qubit utilizes the binding of fluorescent dyes specific to RNA or dsDNA to allow an accurate determination of concentration even in the presence of contaminants.
Bioanalyzer uses a micro-capillary based electrophoretic cell that allows rapid and sensitive quantification, quality control and size analysis of DNA and RNA. . TapeStation is an automated electrophoresis solution for the sample quality control of DNA and RNA samples. Qubit utilizes the binding of fluorescent dyes specific to RNA or dsDNA to allow an accurate determination of concentration even in the presence of contaminants.
Access Protocol
Contact us at single_cell@pasteur.fr