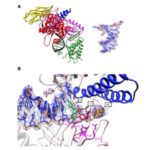
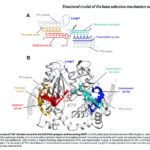
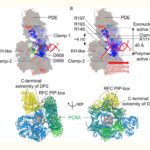
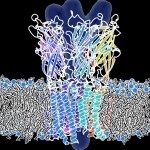
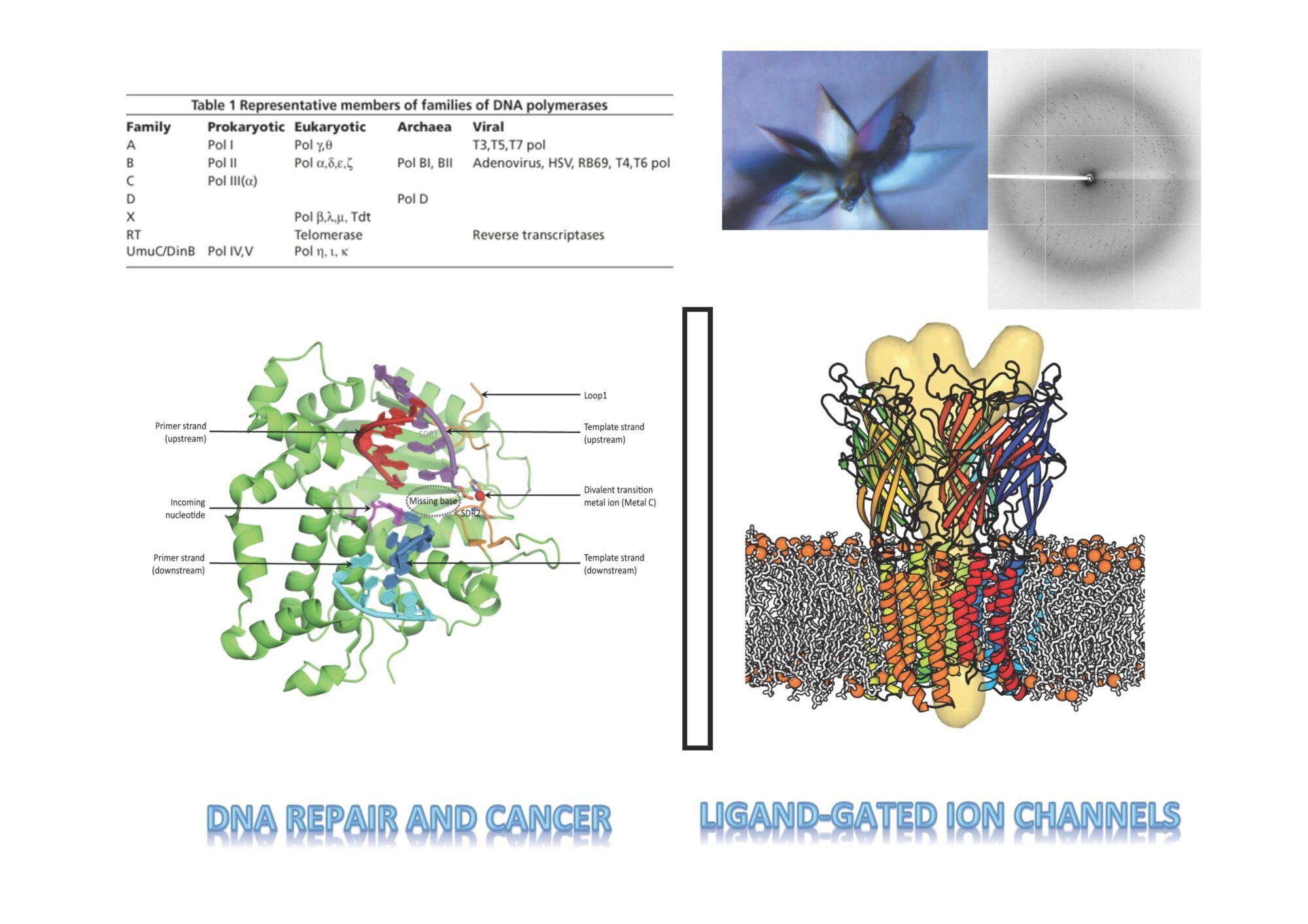
- We use biophysical experimental techniques such as crystallography and cryo-EM to visualize at the atomic level the structure of molecules essential to life, such as DNA polymerases involved in DNA Repair and Cancer and ion channels involved in electric nerve signalling (cell-cell communications in human and bacteria).
- We complement them with molecular dynamics, normal modes dynamics and targeted Langevin dynamics so as to go beyond the essentially static pictures given by these methods and understand the nature of the transition paths between two known forms of the same functional macromolecular assembly.
- In order to be able to predict and interpret the binding properties of macromolecules whose structure is available, we use electrostatics with a realistic model of the solvent and free ions.
- One of our main goals is to design structure-inspired drugs (pharmacology) and re-design active site(s) to make them accept other substrates (synthetic biology).
- We also try to make polymerases evolve by directed evolution to confer them new catalytic properties.
- The sequences of the selected mutants are analyzed by AI methods.
- More details can be found at http://lorentz.dynstr.pasteur.fr (internal users) or at (external users) http://www.dynstr.pasteur.fr
Click to view graph
Connections
About
Groups
Group
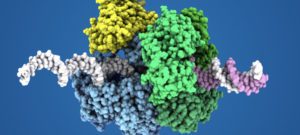
DNA replication group

Ludovic Sauguet
Members
Former Members
2000
2000
Name
Position
2005
2006
Erik Lindahl
Post-doc
2005
2010
Félix Romain
Ingénieur
2005
2008
Jérémie Piton
PhD Student
2005
2008
Cyril Azuara
PhD Student
2008
2014
Jerome Gouge
PhD Student
2008
2012
Hugues Nury
Post-doc
2009
2010
Claudine Mayer
Prof. Paris Cité
2010
2010
Dennis Ptchelkine
Post-doc
2012
2016
Frederic Poitevin
PhD Student
2014
2020
Zaineb Fourati-Kammoun
Post-doc
2015
2020
Frédéric Poitevin
Post-doc
2015
2020
Filippo Romoli
Post-doc
2015
2020
Danièle Sinnaya
Administrative Staff
2015
2020
Irina Volahasina Randrianjatovo-Gbalou
Post-doc
2015
2020
Pierre Raia
PhD Student
2015
2020
Jérôme Loc’h
Post-doc
2015
2020
Haidai Hu
PhD Student
2015
2020
Victoire Prat
Undergraduate Student
2016
2020
Thomas Ybert
Post-doc
2017
2021
Dariusz Czernecki
PhD Student
2018
2022
Clement Madru
Post-doc
2018
2021
Camille Samson
Post-doc
2020
2024
Antonin Nourisson
PhD Student
2021
2024
Rémi Sieskind
Post-doc
2021
2024
Leonardo Betancurt Anzola
PhD Student
2022
2024
Isciane Commenge
Ingénieur
2023
2023
Milena Milovanovic
Master Student
2025
2025
Dariusz Czernecki
Post-doc