About
Bacteria display several mechanisms to adapt to their host-environment and successfully colonise it. One important mechanism of adaptation concerns the regulation of gene expression in response to external signals such as the presence of some scarce nutrients. Binding of a nutrient to a specific membrane transporter triggers a cascade of interactions between several proteins. The signal is then transmitted inside the bacteria where the expression of genes related to the acquisition of the nutrient is activated. In some species, the same cascade regulates the genes associated with bacterial pathogenicity. The understanding of this kind of signalling at the atomic level opens ways towards therapeutic applications. Despite numerous studies, the molecular interactions and the conformational changes involved in this transmembrane signalling are still poorly understood. By using combined approaches of structural biology (NMR, electron microscopy [EM], Small Angle X-ray Scattering [SAXS], biophysical methods) and microbiology, we aim at defining the structural basis of transmembrane signalling pathways at the molecular and atomic levels.
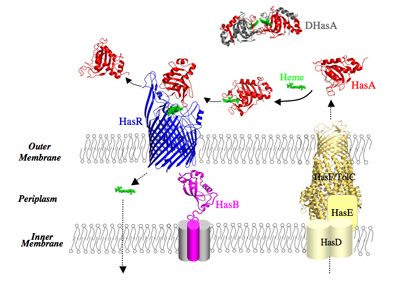
Our model system in a few words: Our model system enables acquisition of heme (iron protoporphyrin IX) as a source of iron. The Has system (Heme Acquisition System) is developed by commensal and pathogenic bacteria. For historical reasons, we study the Has system of Serratia marcescens, a Gram-negative, opportunistic pathogen. In the Has system, heme is acquired through a heme carrier protein HasA (called also hemophore) and an outer membrane receptor HasR. HasA extracts heme from the host’s hemoproteins such as hemoglobin and then delivers it to the HasR receptor. The transport of the heme through HasR and the ejection of the empty HasA are active processes, the energy of which is supplied by the inner membrane HasB complex. HasB is a homologue of TonB protein, essential for acquisition of nutrients (metal complexes, vitamin B12, some carbohydrates) in Gram-negative bacteria.
The expression of genes coding for the proteins of the Has system is inhibited by intracellular iron and activated by the presence and the bioavailability of heme in the external milieu. This external signal is detected by HasR and propagated via a cascade of molecular interactions between different partners. We solved the 3D structure of several proteins of the Has system, free or bound to their partners. All these structural data as well as our in vitro and in vivo data on the molecular interactions involved in different stages of this system make it an excellent model for studying signalling in bacteria.