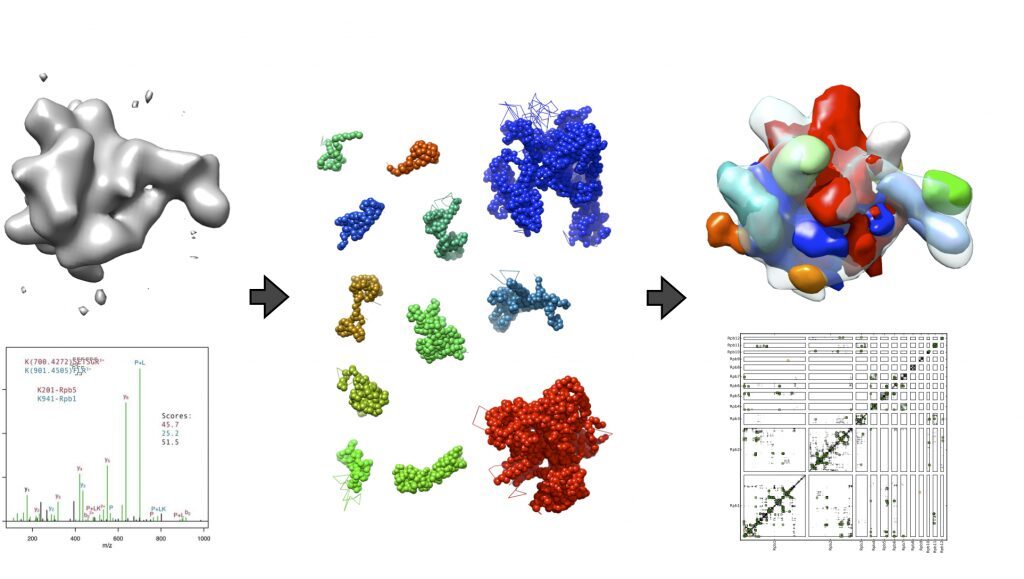
I employ computer modeling methods to study the structure and the dynamics of macromolecular complexes. Such large structures needIntegrative Structure Modeling approaches that encompass the limitations of conventional structure determination methods (crystallography and NMR), by integrating them with unconventional source of information (proteomics, FRET, electron microscopy, statistical inference… ).
To create maximally accurate and optimally precise models, it is crucial to correctly interpret the information contained in experimental data, including its uncertainty. I develop integrative structure modeling approaches that uses Bayes’ theory. These methods allows objective macromolecular structure determination using noisy data derived from heterogeneous samples. These methods are currently being implemented Integrative Modeling Platform (IMP) software. and automatized in the Python Modeling Interface (IMP.pmi) module.
Using computer simulations of coarse-grained polypeptides, I have investigated the kinetics and pathways of protein self-assembly as well as methods to modulate it. I’ve developed coarse-grained models that allow the study of different protein fibrillation scenarios, including the presence of membranes or crowding environment.
I’ve collaborated in the design and synthesis of a peptide with a photoswitchable cross-linker that allows the controlled peptide self-association into amyloid fibrils. Also, I’ve collaborated in the rational design of small compounds that inhibit the formation of Aβ polypeptide aggregates, whose self-assembly is implicated in Alzheimer’s disease. With the goal of designing a modular repeat protein as a specific binder for peptides, I’ve developed a molecular dynamics approach to predict mutations that stabilize the protein from a molten globule to a cooperative folder.