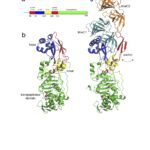
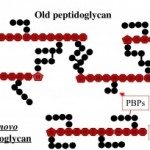
The cell envelope of Gram-negative bacteria is a complex structure composed of two membranes and a cell wall. The cell wall determines cell shape and protects the cell from bursting due to osmotic pressure. Depending on the growth conditions and the environment bacteria can undergo shape transitions, both on short-term and evolutionary time scale. During cell elongation and cell division, new building blocks are incorporated into the cell wall and old material recycled to allow cells to grow and divide. This process is tightly coordinated and directly linked to insertion of proteins into the outer membrane, their covalent interaction with the cell wall, and synthesis of surface lipopolysaccharides. The activity of essential membrane protein complexes involved in these events depend on lipoproteins. Lipoproteins are modified by fatty acids derived from membrane phospholipids by a sequential action of three integral cytoplasmic membrane enzymes. They are involved in cell wall synthesis directly as cell wall hydrolases or through stimulation of the enzymatic activity of penicillin-binding proteins. They are themselves virulence factors and interact with host Toll-like receptors via their lipid moiety. Over the years, we reported findings on the lipoprotein modification process by determining the membrane topology of the enzymes, their enzymatic activity and substrate specificity, and the molecular mechanism of the reactions. We also investigated the effect on cell morphology when lipoprotein modification is perturbed to understand the basis of its essentiality for bacterial viability.
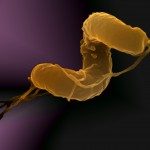
We are interested in understanding how the cell envelope contributes to cell shape transitions and how shape influences virulence. We use Helicobacter pylori as model organism, which undergoes a morphological transition from spiral-rod to coccoid as a mechanism to escape the innate immune response. We focus on the connection, and its regulation, between outer membrane lipoproteins and the cell wall. The outcome of this fundamental question contributes to the identification of vaccine candidates and the development of novel antibacterial agents.