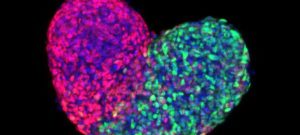
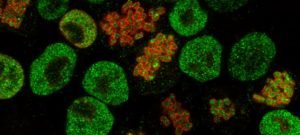
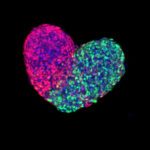
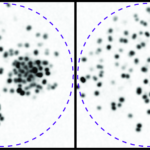
The Unit for the Physics of Biological Function studies the basic physical principles that govern the existence of multicellular life. A core focus of the lab is to understand biological development–the complex process through which an organism grows from a single cell into a differentiated, multicellular organism–from a physics perspective. As such, we formulate and experimentally validate quantitative models that describe how individual cells interact and organize in order to generate complex life forms.
Our main interests lie in:
- multicellular pattern formation
- transcriptional regulation in the context of development
- molecular limits to biochemical sensing
- emergence of collective behaviors in multicellular systems.
Our main tools consist in:
- live and super-resolution microscopy
- genome editing and genetics
- image analysis and quantitative modeling
- instrument design and development.
Work with us
We are seeking postdoc candidates with interests in the broader fields of biophysics and/or cell and developmental biology. In particular, physicists or engineers with expertise in a subset of nonlinear and ultrafast optics, super-resolution imaging, instrumentation (design and implementation), photomanipulation, optogenetics, advanced image analysis, statistics, live imaging of stem cells, tissues, or embryos; biologists or computer scientists with expertise in a subset of multicellular development, gene regulation and epigenetics, cloning and transfection, genetics, live imaging.
Specific topics (in progress…):
- precision and reproducibility in early mammalian development
- size control in mammalian development
- super-resolution imaging of subnuclear structures and single molecules in vivo
- optogenetic pathway activation and perturbation
- information transmission in developmental pathways
- …
Open positions
We are currently looking to fill positions for postdoctoral training and are looking for doctoral and master students to join the lab. They center around three themes that we want to push forward over the next two to three years:
1. At the level of a genetic network, extract global properties and design principles from expression level measurements and analysis.
2. At the molecular level, develop mathematical models underlying the fundamental mechanisms of transcriptional regulation. We test these using single molecule and live measurements of the transcriptional output, and perturbative experiments using genetics and opto-genetics.
3. At the level of the dynamics of the DNA polymer, link the nuclear architecture with actual transcriptional activity in terms of multiple enhancers recruiting the same promoter in a given cell.
Experimental and theoretical physicists (preferentially with experience in biological and/or soft-matter physics), engineers and materials scientists with experience in biology, and biologists interested and experienced in quantitative approaches are strongly encouraged to apply. Outstanding applicants in other fields (mathematics, computer science, chemistry, etc.) will also be considered. Experience with microscopy and image analysis is a plus but not necessary.
Applicants should email me their CV and a brief description of research interests, as well as their motivation to join the lab.