Welcome to the laboratory “Membrane Biochemistry and Transport”!
Have you ever wondered how human cells get rid of waste? If yes, we have some answers and exciting research projects that aim to reveal the fundamental principles of autophagy, the most versatile cellular recycling pathway.
What is autophagy?
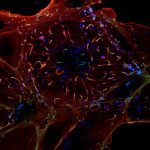
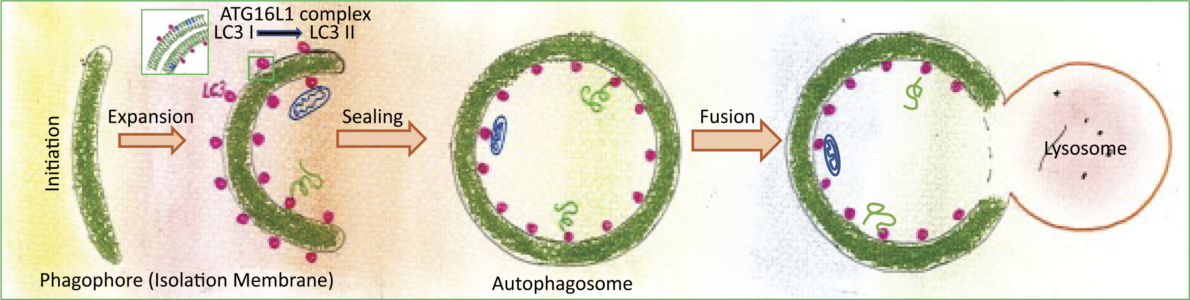
Autophagy is a fascinating cellular degradation pathway that selects cytoplasmic material, sequesters it in autophagosomes and transports it to lysosomes for degradation. Autophagic cargo, including organelles, protein aggregates, ribosomes and multienzyme complexes, is captured by a membrane cisterna, termed phagophore. The membrane expands around its cargo to engulf it entirely and sealing of the membrane gives rise to an autophagosome, which is a “transport container” that delivers its content to lysosomes.
Why do we study autophagy?
The importance of autophagy for the homeostasis and stress resistance of human cells cannot be appreciated enough. Autophagy takes a central position in cellular catabolism and is interconnected with many other cellular pathways. Reduced autophagic activity or dysfunctions are contributing to the onset of many human diseases, including cancer, neurodegeneration, metabolic and immune diseases. Such a decline of autophagy occurs during aging, leading to a higher prevalence of neurogenerative diseases and cancer in the older population. A directed and precise manipulation of the autophagic activity offers so far unexplored options to treat those diseases, notably neurodegenerative diseases for which no efficient treatment exists.
How do we study autophagy?
We are combining two different approaches to investigate autophagy. The first aims to reconstitute the formation of autophagosomes in the test tube from purified components. We use model membranes which are assembled from synthetic lipids and purified proteins, which we express recombinantly using E.coli, insect, or human cells.
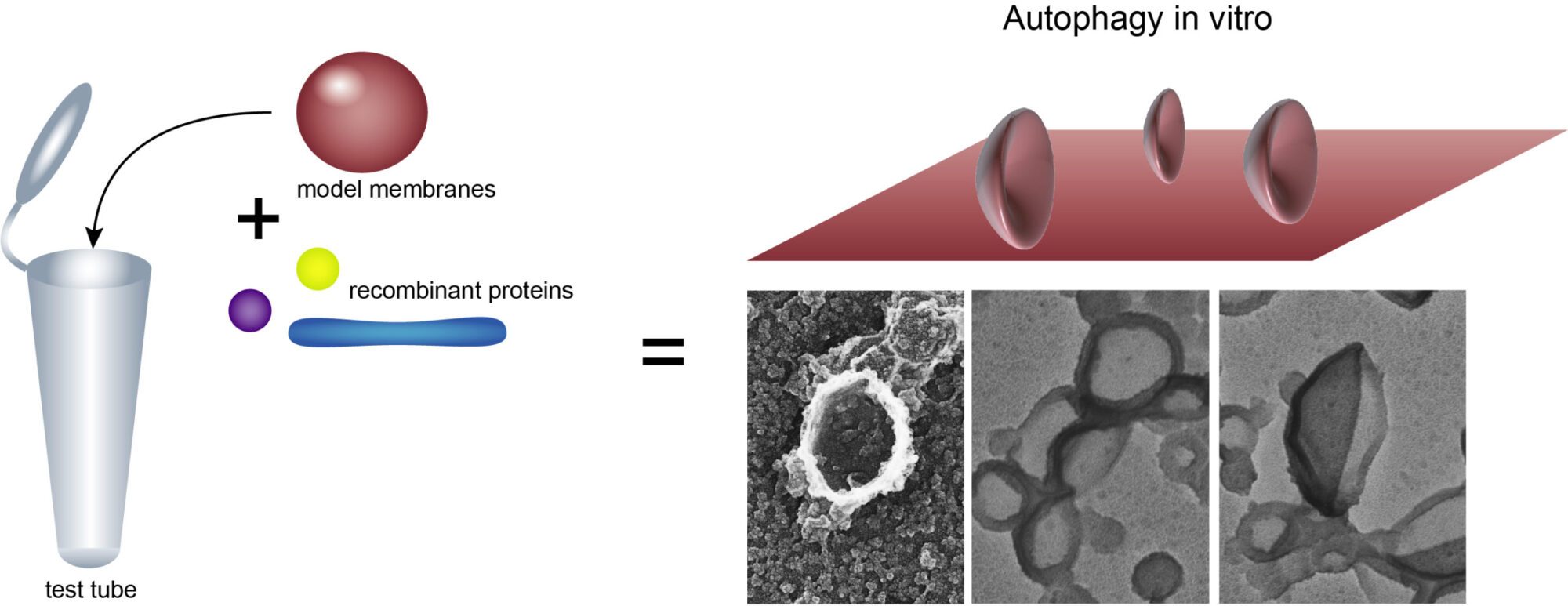
The second approach combines cell biology, biochemistry, biophysical analysis and structural studies in cultured cells. Here, we uncouple parts of the autophagy machinery from the pathway by redirecting them to other membranes, for example the plasma membrane or organellar membranes. This “in vivo” reconstitution approach allows us to study the activity of components of the autophagy machinery and their molecular function independently of canonical autophagy. Therefore, we express fluorescent labeled proteins and study their interaction on membranes by e.g. fluorescence lifetime imaging. The hallmark of autophagy is the formation of phagophores and autophagosomes, which depends on massive remodeling of cellular membranes. In order to correlate protein function with such membrane deformations, we apply electron microscopy techniques including correlative light electron microscopy, focused ion beam – scanning electron microscopy and cryo-electron tomography.
Autophagy and neurodegeneration
We recently identified a specialized autophagy pathway that degrades protein aggregates in neural cells. We found that autophagosomes that transport protein aggregates to lysosomes are decorated with LC3C, a small ubiquitin like protein which belongs to the ATG8 protein family. LC3C is covalently attached to lipids of the phagophore membrane and provides identity to such autophagosomes. The destination of LC3C labeled autophagosomes is defined by TECPR1, a protein that is found on a subtype of lysosomes that contain the lipid PtdIns(4)P in their membrane. TECPR1 selectively interacts with LC3C to recruit protein aggregate containing autophagosomes. Targeting TECPR1 to endosomes leads to a missorting of LC3C autophagosomes, which are no longer delivered to lysosomes for degradation but accumulate at endosomes instead.
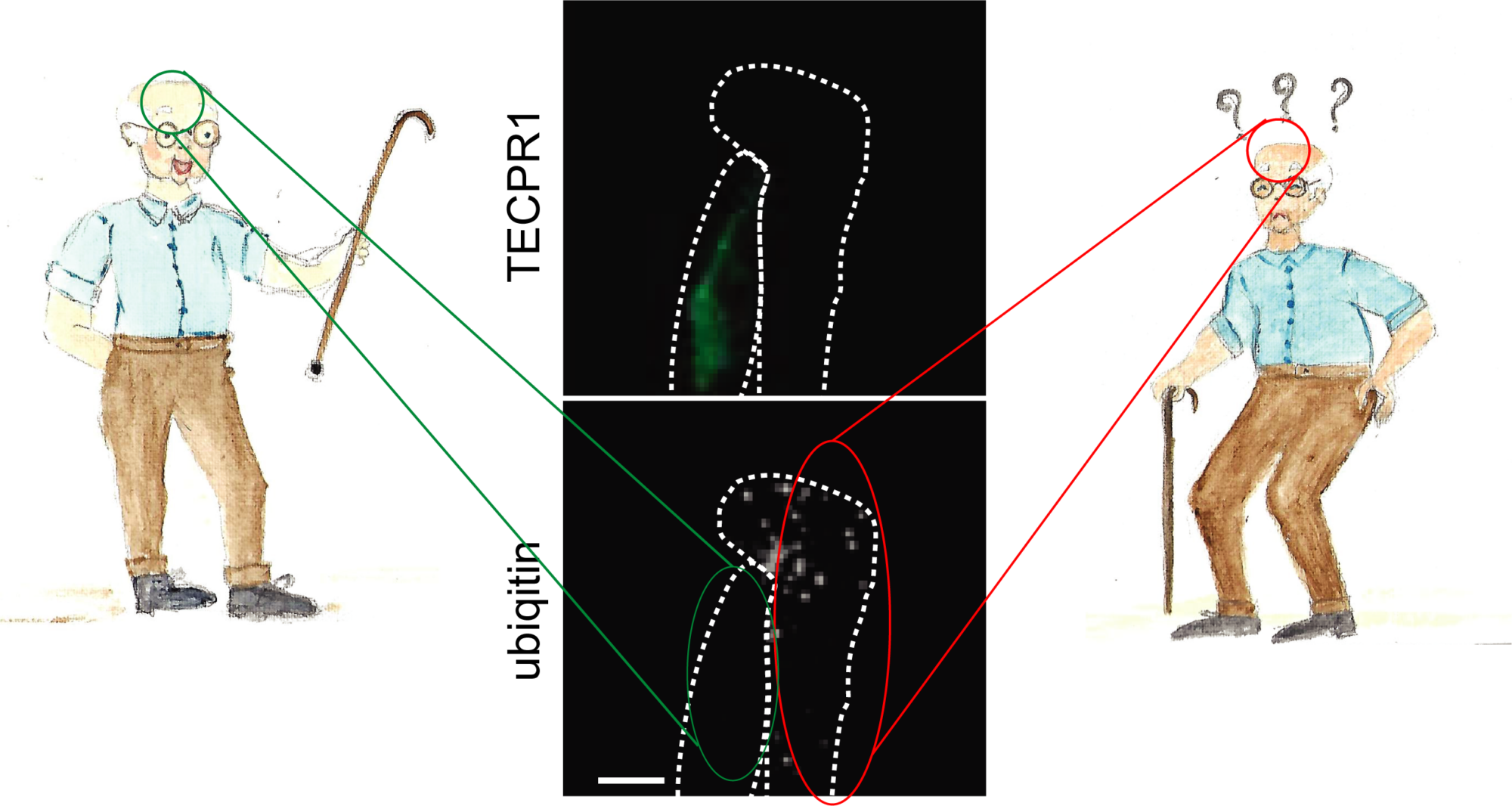
Right: The activity of autophagy decline with age, and in some older people, protein aggregates accumulate in neurons of the brain, leading to neurodegeneration.
We found that impaired autophagy, which entails an accumulation of protein aggregates, can be reverted. If protein levels of TECPR1 in neural cells are restored, the autophagic activity in these cells is augmented. This leads to an enhanced clearance of protein aggregates, protecting neural cells from cytotoxicity and cell death. Many neurodegenerative diseases are caused by an accumulation of proteins including alpha-synuclein and Tau in the case of Alzheimer’s and Parkinson’s diseases. Our findings demonstrate that neurodegeneration can be treated by selectively enhancing TECPR1 levels.
If you want to lean more about this project, you can click on this link: Paper “TECPR1 promotes aggrephagy by direct recruitment of LC3C autophagosomes to lysosomes”
Nonselective autophagy and stress response
Autophagy does not only degrade cytoplasmic waste, it also degrades bulk cytoplasm in cells that encounter cytotoxic stress or in response to starvation. But there is a major difference between selective autophagy of cytoplasmic cargo and nonselective autophagy of bulk cytoplasm. In selective autophagy, the membrane wraps around cargo. The shape of the membrane is thus defined by the shape of cargo. In nonselective autophagy, bulk cytoplasm, which is a viscous solution, needs to be captured by a membrane that behaves like a two-dimensional liquid. Thus, nonselective autophagy requires that phagophores are shaped into free standing membrane cups, which expand without templating cargo to enclose portions of cytoplasm. How this can be achieved, remained a longstanding question. We recently found that another member of the ATG8 protein family, LC3B and its E3-ligase complex, which catalyzes the conjugation of LC3B to membranes, play a key role in the process. Both assemble into two-dimensional scaffolds that deform flat donor membranes into membrane cups. By reconstituting this process on model membrane in vitro and at the plasma membrane of cells, we revealed unprecedented insights into the molecular mechanism of this process and identified a subunit of the E3 ligase complex, ATG16L1, as a key driver of cup formation.
If you are interested to lean more about this project, you can read the full story at: Preprint “Phagophore formation by the autophagy conjugation machinery”
Team Building at Parc Asterix June 2024:
Christmas Party 2022: Bubble foot
Social Activities 2019: Birthday Party for Thomas

PhD celebration Peter

2018: World Soccer Championships 2018

Birthday Cake for Thomas

Birthday Cake for Jagan

Birthday Cake for Peter

2017: Christmas Party