About
To understand protein functions and molecular network organisation, it is important to determine the localisation of proteins at subcellular level. The in cellulo probing of molecular assemblies is usually performed with microscopy techniques which visualise the localisations of molecules. The analysis of the spatial proximity between different molecules of interest is traditionally achieved by labelling proteins with different fluorophores (in general red and green fluorophores), and quantifying their spatial co-localisation. Traditional co-localisation index are based on signal correlation (e.g. Pearson Correlation Coefficient) and overlap (e.g., Manders Overlap Coefficient) and thus depend on the size of the point spread function (PSF) of the microscope. Moreover, these methods cannot be directly used for single-molecule localisation microscopy where the nanometre scale localisation of molecules is directly estimated from the sequential activation of blinking molecules. Another important issue in any co-localisation analysis is the topographical organisation of molecules within the cell that can frequently bias the interpretation of co-localisation coefficients. Indeed, confined molecules can strongly overlap or be very close (localisations) even if randomly distributed.
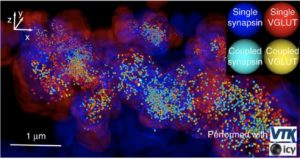
To robustly and statistically characterize the spatial coupling between objects using traditional or super-resolved microscope, we are developing a method and software named Statistical Object Distance Analysis (SODA). Contrary to previous co-localisation methods, SODA allows to map statistically coupled objects within the cell, i.e., to compute a coupling probability for each single pair of objects. As SODA uses a marked-point process framework, it does not depend on the PSF characteristics and is robust to noise. In addition, by analysing the morphology (size, intensity and shape) and the distance separating coupled molecules, it provides, at a population level, a detailed and exhaustive description of the molecular assemblies.