The CNR for enterohaemorrhagic Escherichia coli, Shigella and Salmonella (CNR-ESS) is in charge of the microbiological surveillance of these three pathogens in humans in France through a network of > 1000 clinical laboratories. This centre deals with more than 10,000 isolates per year and has been working in very close collaboration with the epidemiologists on the detection and investigation of outbreaks using a wide range of molecular tools (from pulsed field gel electrophoresis to whole genome sequencing). In 2008, it was estimated that the CNR-ESS surveillance system detected 66% of laboratory-confirmed human Salmonella infections.
Beyond routine activities in surveillance and outbreak investigations, the applied research activities of the CNR is in accordance with the terms of reference issued by the Ministry of Health for the current mandate.
- Development of new typing and subtyping tools with the long-term goal of replacing serotyping or subtyping by sequence-based typing methods based on MLST, CRISPR or single-nucleotide polymorphisms,
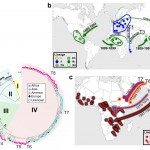
- Identification, characterization and dynamics of transmission of antibiotic-resistant bacterial populations, like the highly-drug resistant enterica serotype Kentucky ST198 strain which spread globally, or detection of any new emergent mechanisms of antibiotic resistance
- Collaboration with biostatisticians for developing new Salmonella surveillance modelisation tools.