HIV was discovered in 1983 and the reverse transcription (RT) process that characterizes all retroviruses in 1970. However, new notions on the RT were deciphered only recently.
Of note, our group has played a major role in revealing new spatiotemporal aspects of RT, uncoating (loss of the viral capsid shell) and the topology of HIV pre-integration, maturation and integration. Inside the host nucleus, newly synthesized viral DNA (vDNA) was found in HIV-1-induced cleavage and polyadenylation specific factor 6 (CPSF6) / serine/arginine-rich splicing factor (SC35) organelles. We called these viral/host structures HIV-1 membraneless organelles (HIV-1 MLOs). The aim of our research is untangling the mechanism underlying HIV-1 MLO biogenesis and their role in viral retrotranscription, replication and latency.
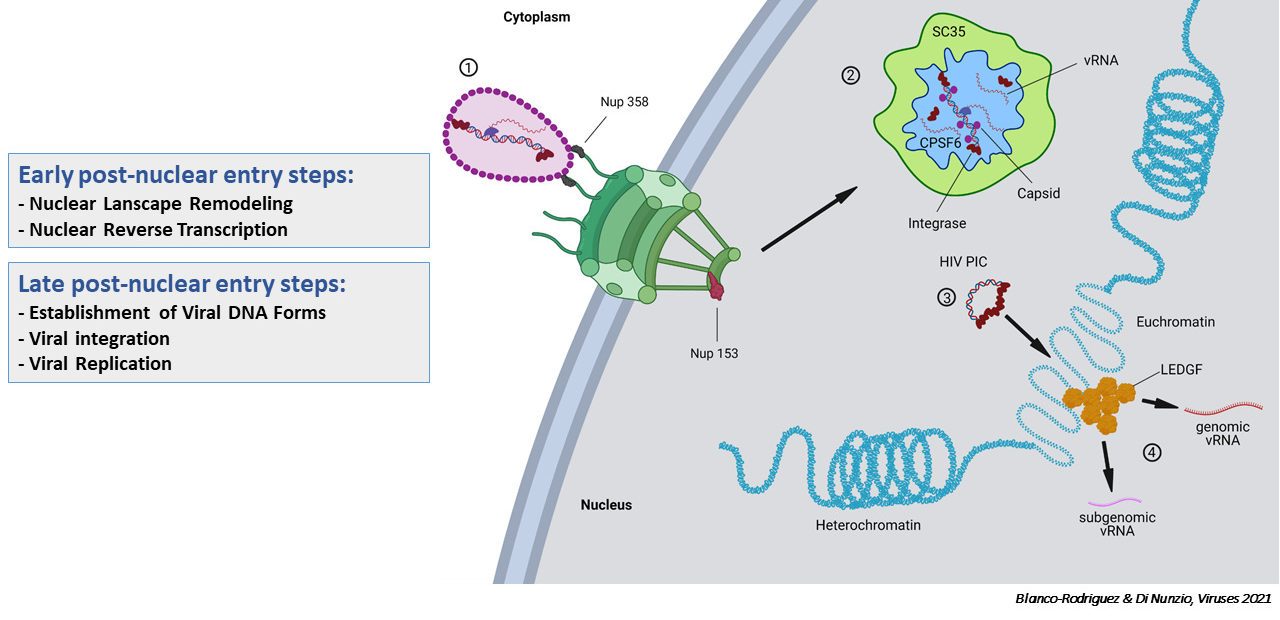
We propose a bipartite model of HIV post-nuclear entry steps:
Next steps include in vivo studies.
In particular, we aim at elucidating mechanism/s in common among divergent viruses that interact with host components inside MLOs and sabotage cellular compartments to their advantage.
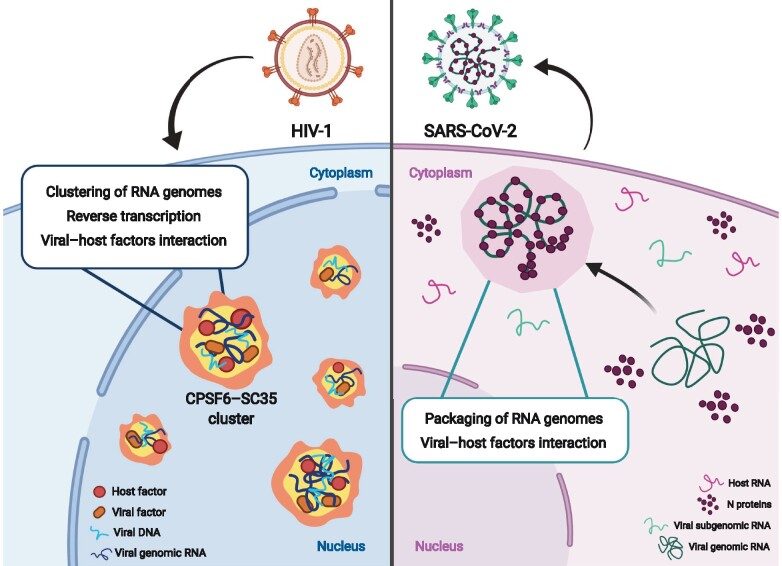
MLOs induced by HIV-1 and SARS-CoV-2 to replicate from Scoca & Di Nunzio JMCB 2021. Left: HIV-1 infection prompts the formation of nuclear MLOs enriched with host factors, such as CPSF6 and SC35, and in viral components, such as vRNA, vDNA, capsid, and integrase. Right: SARS-CoV-2 N protein forms condensates in the cytoplasm to recruit exclusively intact vRNA genome against subgenomic vRNAs or host RNAs . Cartoon created with BioRender.com.