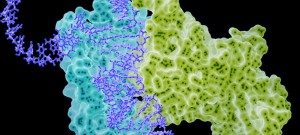
Our focus as the chemoinformatics and proteochemometrics group is to discover and analyze substances that can modulate macromolecular interactions, with a particular emphasis on protein-protein interactions (PPIs), utilizing computational methods. We combine chemoinformatics and structural bioinformatics techniques to comprehend the structural characteristics of binding sites situated at the core of PPI interfaces, as well as the optimal physicochemical properties required for small molecules to modulate these interactions.
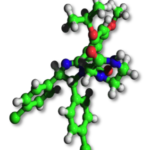
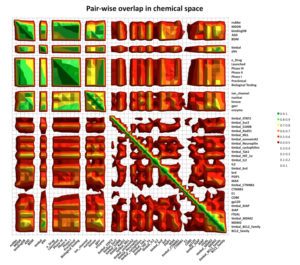
A significant aspect of our work involves studying successful examples of pharmacological modulation of PPIs to gain insights into the chemical space of PPI inhibitors. To facilitate this, we have initiated the iPPI-DB project (http://ippidb.pasteur.fr/), a dedicated database focused on small molecule PPI modulators. This database serves as a valuable resource for pharmacological data, enabling us to derive trends about the chemical space of PPIs using machine learning techniques.
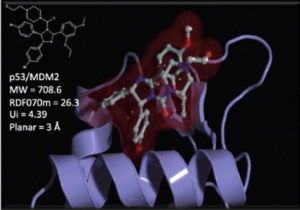
Our expertise also extends to designing project-specific chemoinformatics-based chemical libraries. These libraries are customized to cater to specific research projects and can be utilized for virtual ligand screening (VLS) within our team or for conducting high-throughput screening (HTS) campaigns. In pursuit of this objective, we have collaborated with the group of Xavier Morelli at CRCM in Marseille, utilizing the valuable data from iPPI-DB. This collaboration has allowed us to jointly develop and co-design the Fr-PPIChem library, incorporating insights from the iPPI-DB data.
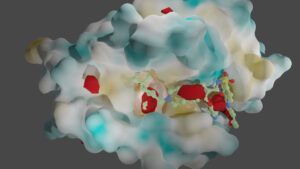
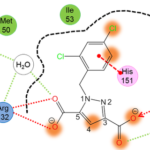
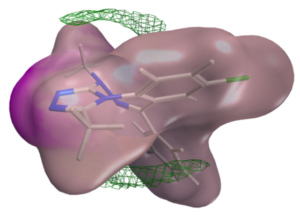
Another essential aspect of our project is to investigate the properties of PPI interfaces themselves, as they influence binding modality, privileged chemotypes, and protein motifs favoring ligand binding, all of which can be effectively utilized by machine learning tools. We are mapping the entire PPI pocketome, encompassing all the binding sites within PPIs, to assess the suitability of these binding pockets as targets for small molecule ligands. By combining information about target proteins and the chemical space of PPIs, our aim is to determine which chemotypes should be associated with specific types of PPI targets. This endeavor seeks to facilitate the identification of high-quality chemical probes for PPI targets and other macromolecular interactions, employing complementary chemical biology approaches.
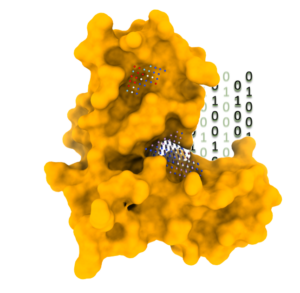
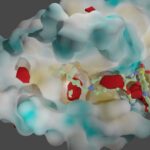
Furthermore, we utilize advanced computational techniques such as 3D fully convolutional neural networks, including the U-Net model, to detect and analyze functional binding sites within protein-protein interactions. By harnessing the power of deep learning and convolutional neural networks, we can accurately identify critical regions involved in protein-protein interactions.
To facilitate our research in collaboration with Dr Guillaume Bouvier, we have actively coordinated the development of InDeep, a platform that allows researchers to access and leverage the capabilities of 3D fully convolutional neural networks for detecting functional binding sites within protein-protein interactions. InDeep serves as an invaluable resource, offering a user-friendly interface accessible at https://indeep-net.gpu.pasteur.cloud/. Our goal is to foster collaborations and empower the scientific community to explore and comprehend the intricate aspects of protein-protein interactions at a functional level through this accessible tool.