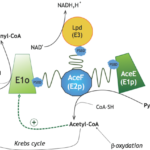
Our group studies the composition and molecular architecture of protein complexes involved in central metabolism in Actinobacteria, and the signaling pathways and/or allosteric mechanisms that contribute to their regulation. The objective is to shed light on the mechanisms that underlie the metabolic plasticity of the members of this phylum. Our experimental approaches involve the integrative structural biology of large metabolic complexes from the non-pathogenic species Corynebacterium glutamicum, a well-known workhorse for the biotech industry, but we are also interested in structural enzymology for drug design purposes, especially for targets from Mycobacterium tuberculosis. More information about our current activities can be found on the SUPERCPLX and METACTINO project pages.
Click to view graph
Connections
Members
Former Members
2000
2000
Name
Position
2018
2022
Lu Yang
PhD student
2019
2021
Monika Zahorszka
Visiting PhD Student
2020
2021
Alexandra Boyko
PhD student
2014
2020
Eduardo Bruch
Postdoc
2020
2020
Vasily Aleshin
Visiting PhD Student
2018
2019
Matteo Mori
Visiting PhD Student
2017
2018
Pierre Vilela
Graduate Student
2014
2017
Isabelle Miras
Technician
Projects
Fundings
Featured publications
-
2021Actinobacteria challenge the paradigm: A unique protein architecture for a well-known, central metabolic complex., Proc Natl Acad Sci U S A 2021 Nov; 118(48): .
-
2021A Tetratricopeptide Repeat Scaffold Couples Signal Detection to OdhI Phosphorylation in Metabolic Control by the Protein Kinase PknG., mBio 2021 Oct; 12(5): e0171721.
-
2019Structural insights into the functional versatility of an FHA domain protein in mycobacterial signaling., Sci Signal 2019 05; 12(580): .
-
2019Novel mechanistic insights into physiological signaling pathways mediated by mycobacterial Ser/Thr protein kinases., Genes Immun 2019 05; 20(5): 383-393.
-
2019New substrates and interactors of the mycobacterial Serine/Threonine protein kinase PknG identified by a tailored interactomic approach., J Proteomics 2019 Feb; 192(): 321-333.
-
2017PknG senses amino acid availability to control metabolism and virulence of Mycobacterium tuberculosis., PLoS Pathog 2017 May; 13(5): e1006399.
-
2017The crystal structure of PknI from Mycobacterium tuberculosis shows an inactive, pseudokinase-like conformation., FEBS J 2017 02; 284(4): 602-614.
-
2015Thiophenecarboxamide Derivatives Activated by EthA Kill Mycobacterium tuberculosis by Inhibiting the CTP Synthetase PyrG., Chem Biol 2015 Jul; 22(7): 917-27.
-
2015The crystal structure of the catalytic domain of the ser/thr kinase PknA from M. tuberculosis shows an Src-like autoinhibited conformation., Proteins 2015 May; 83(5): 982-8.
-
2014A dual conformation of the post-decarboxylation intermediate is associated with distinct enzyme states in mycobacterial KGD (α-ketoglutarate decarboxylase)., Biochem J 2014 Feb; 457(3): 425-34.
-
2013GarA is an essential regulator of metabolism in Mycobacterium tuberculosis., Mol Microbiol 2013 Oct; 90(2): 356-66.
-
2011Functional plasticity and allosteric regulation of α-ketoglutarate decarboxylase in central mycobacterial metabolism., Chem Biol 2011 Aug; 18(8): 1011-20.
-
2008Regulation of glutamate metabolism by protein kinases in mycobacteria., Mol Microbiol 2008 Dec; 70(6): 1408-23.
-
+View full list of featured publications
-
+View full list of publications
Contact
Phone: +33-1-45688608 Email: marco.bellinzoni@pasteur.fr Address 25-28 Rue du Docteur Roux 75015, Paris France