Link to Pubmed [PMID] – 27903652
Link to DOI – 10.1074/jbc.M116.759498
J Biol Chem 2017 Jan; 292(1): 328-338
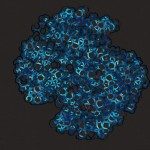
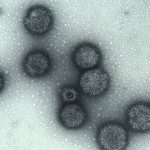
Members of a group of multimeric secretion pores that assemble independently of any known membrane-embedded insertase in Gram-negative bacteria fold into a prepore before membrane-insertion occurs. The mechanisms and the energetics that drive the folding of these proteins are poorly understood. Here, equilibrium unfolding and hydrogen/deuterium exchange monitored by mass spectrometry indicated that a loss of 4-5 kJ/mol/protomer in the N3 domain that is peripheral to the membrane-spanning C domain in the dodecameric secretin PulD, the founding member of this class, prevents pore formation by destabilizing the prepore into a poorly structured dodecamer as visualized by electron microscopy. Formation of native PulD-multimers by mixing protomers that differ in N3 domain stability, suggested that the N3 domain forms a thermodynamic seal onto the prepore. This highlights the role of modest free energy changes in the folding of pre-integration forms of a hyperstable outer membrane complex and reveals a key driving force for assembly independently of the β-barrel assembly machinery.