
Klebsiella pneumoniae [17]

Group : Molecular microbial evolution & ecology
Olaya Rendueles-Garcia

Virgile Andreani
Physicist and computer scientist by training, I develop quantitative population models of antibiotic resistance, which I calibrate using optimal experimental design. I then exploit them to engineer optimal treatment strategies.
Imaging and pathophysiology of Klebsiella pneumoniae and Klebsiella rhinoscleromatis infections
Régis Tournebize
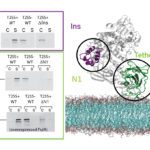
Type II protein secretion mechanism
Olivera Francetic
Genomic and nomenclature databases of pathogenic bacterial strains
Sylvain Brisse
Population biology of the bacterial pathogen Klebsiella pneumoniae
Sylvain Brisse
Genomic taxonomy of bacterial strains: universal nomenclatures for epidemiology and population biology
Sylvain Brisse

FiberSpace
Olivera Francetic

Lulla Opatowski
Epidemiology and Modelling of Antibacterial Evasion (EMAE)
Lulla Opatowski is professor in Mathematical Epidemiology (University of Versailles Saint Quentin – UVSQ). My main research interests are in bacterial resistance to antimicrobials (including antibiotics and vaccines) and host-pathogen-pathogen interactions, through the combination […]

Didier Guillemot
Epidemiology and Modelling of Antibacterial Evasion (EMAE)

Microbial Evolutionary Genomics
Eduardo Rocha
Epidemiology and Modelling of Antibacterial Evasion (EMAE)
Didier Guillemot

Jean-Marc Cavaillon
Jean-Marc Cavaillon is honorary professor of Institut Pasteur where he made his whole career after his post-doctoral training at the University of Toronto (Canada). He has been head of a research Unit “Cytokines & […]

Olivera Francetic
Macromolecular Systems

Eduardo Rocha
UMR3525 CNRS/Institut Pasteur – Genetics of genomes
My work focuses on the interplay between the organisation of genomes arising by the chromosomal interactions of cellular processes and the recombination processes generating adaptive genetic variation. Genetic elements tend to be organized in […]