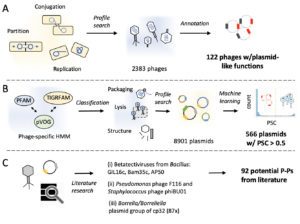
Plasmids and temperate phages are key contributors to bacterial evolution, but are usually regarded as very distinct. Some elements, termed phage–plasmids, are known to be both plasmids and phages. The number, distribution, relatedness and characteristics of these phage–plasmids are poorly known. Here, we screened for these elements among thousands of genomes and identified 780 phage–plasmids across very diverse bacterial phyla. We grouped 92% of them by similarity of gene repertoires to eight defined groups and 18 other broader communities of elements. The existence of these large groups suggests that phage–plasmids are ancient. Their gene repertoires are large, the average element is larger than an average phage or plasmid, and they include slightly more homologs to phages than to plasmids. We analyzed the genetic diversity and organization of each group of phage–plasmids and found the key phage genes to be conserved and co-localized within distinct groups, whereas genes with homologs in plasmids are much more variable and include most accessory genes. Phage–plasmids are a sizeable fraction of the sequenced plasmids (∼7%) and phages (∼5%), and could have key roles in bridging the genetic divide between phages and other mobile genetic elements.
© Research
News