Présentation
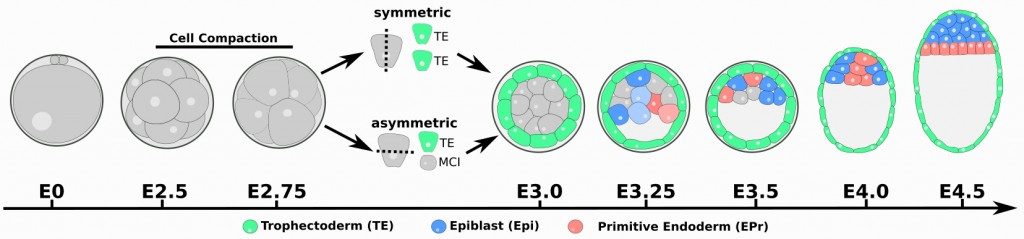
The mammalian preimplantation embryo is a model of choice to study these processes and derivation of stem cells from early embryos has generated a lot of interest for this period of development. Mammalian development starts with a period when embryo evolves freely within the maternal reproductive tracts to reach its final destination, the uterus, where it will implant into the uterine wall after 4.5 days of development (E4.5) in the mouse. During this journey, several critical events will happen such as parental genome reprogramming, zygotic genome activation, polarity acquisition… This period is also allocated to the preparation of the embryo for implantation with the specification and segregation of embryonic and extraembryonic cell lineages. At the end of the preimplantation period, the blastocyst comprises three molecularly and spatially distinct cell lineages. The trophectoderm (TE), a single epithelial layer encapsulating the blastocoel cavity and an inner cell mass (ICM) composed of the pluripotent epiblast (EPI) and an epithelial layer of primitive endoderm (PrE) in contact to the cavity. EPI will essentially contribute to the embryo proper while TE will contribute to the fetal portion of the placenta and PrE to the yolks sac layers as well as part of the endoderm of the fetus.Two specification steps are necessary for the specification of lineages: TE and Inner Cell Mass (ICM) are specified between E2.5 and E3.0 (1). Epi and PrE lineages are established within the ICM between E3.0 and E3.75 (2).
Mouse preimplantation development
Our lab is interested in the molecular mechanisms driving the divergence between EPI and PrE cell fates within the ICM with a particular emphasis on the role of signalling molecules and key-lineage transcription factors. To dissect these processes, we are combining live imaging tools to genetic and pharmacological approaches both in vivo and in stem cell lines representative to these lineages. Recently, compilation of data obtained in vivo in a mathematical model allows to conclude that the core regulatory network (CRN), composed of NANOG, FGF4, GATA6 and FGFR2, is necessary and sufficient to carry out ICM cell specification into Epi and PrE (3). However, what are the factors that act upstream the CRN and how specification is initiated remains largely unknown. Our aim is to elucidate the molecular mechanisms that govern Epi/PrE cell specification and differentiation. This project will be performed in collaboration with the group of Claire Chazaud (GReD Institute, Clermont-Ferrand) in the frame of the ANR 2014 PrEpiSpec project.
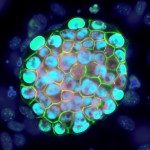
® Sylvain Bessonnard
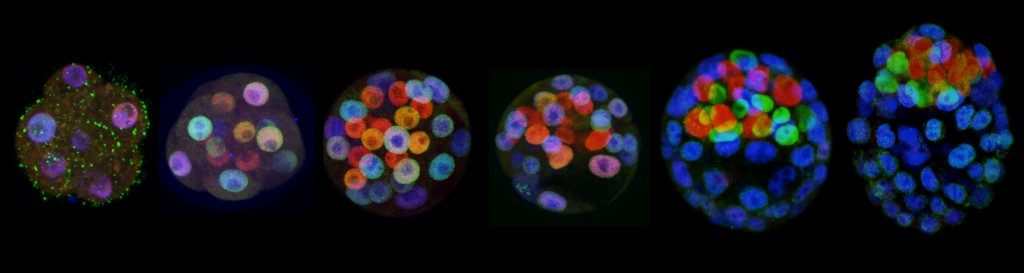
Progressive segregation of Epi and PrE lineages visualized by NANOG (red) and GATA6 (green) immunodetetection
- Sasaki, Seminars in Cell and Developmental Biology, 1–8 (2015).
- J. Artus, Cell. Mol. Life Sci. 71, 3327–3338 (2014).
- S. Bessonnard, Development. 141, 3637–3648 (2014).