Understanding the biology of infection requires a quantitative perspective on how living systems sustain stable dynamical processes and how these are perturbed by microbial challenges. Biological functions are not static but emerge from dynamic, multiscale interactions among agents—ranging from molecules driving cellular processes, to cells coordinating within tissues, to neurons shaping organismal behavior. Disentangling these interactions, and elucidating how pathogens or environmental stressors disrupt them, is essential for uncovering fundamental biological principles and identifying new therapeutic opportunities.
Our goal is to establish an integrative framework built around three methodological axes:
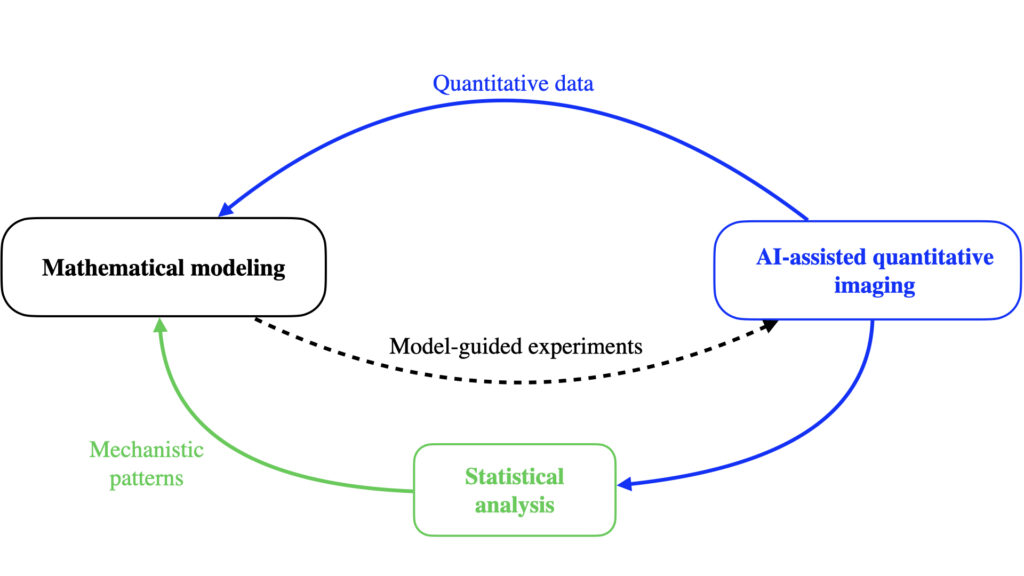
– Mathematical modeling of dynamic networks, to explain how stable biological processes emerge from local interactions and to predict how they are perturbed.
– Statistical analysis of spatial and temporal relations between agents, to extract mechanistic insights from complex observational datasets.
– AI-assisted image quantification, to transform high-dimensional imaging data into interpretable features that inform models and analyses.
We will apply these approaches to two complementary systems that provide a comparative perspective on biological processes across scales—from single molecules to whole organisms—and how their stability is perturbed by microbial influence.
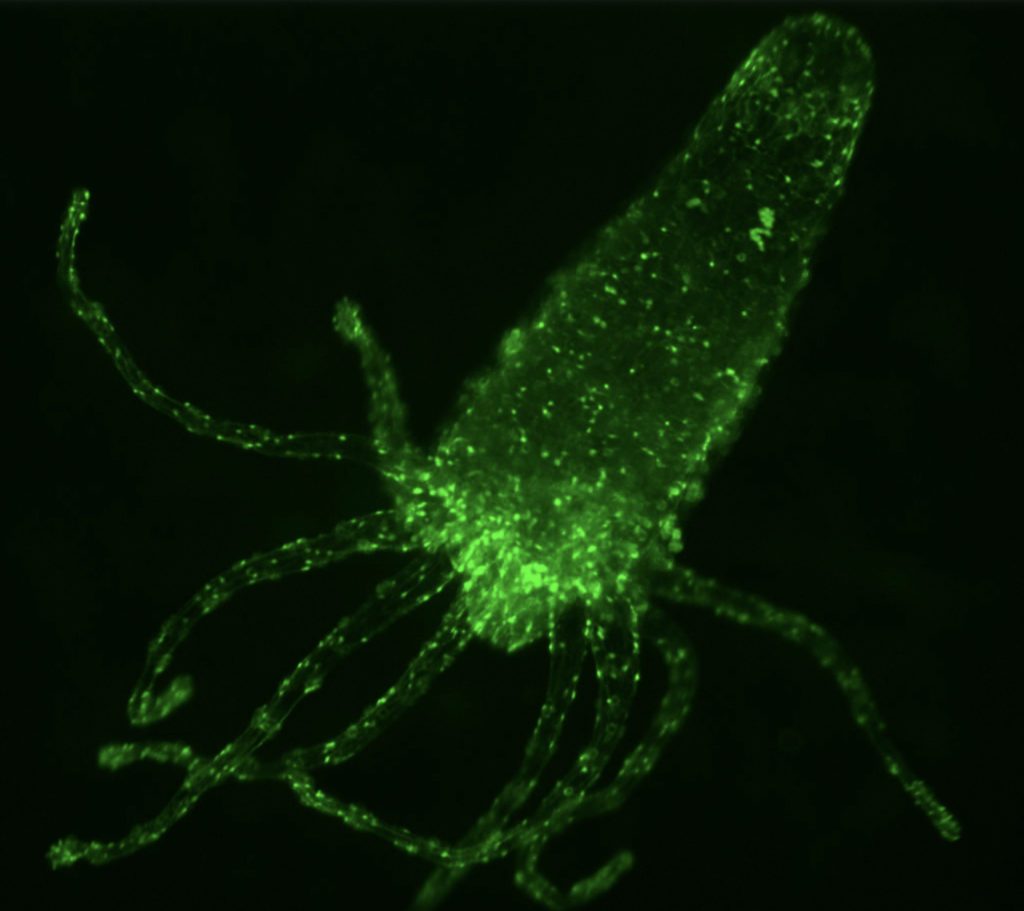
Hydra as a paradigm for neuronal homeostasis.
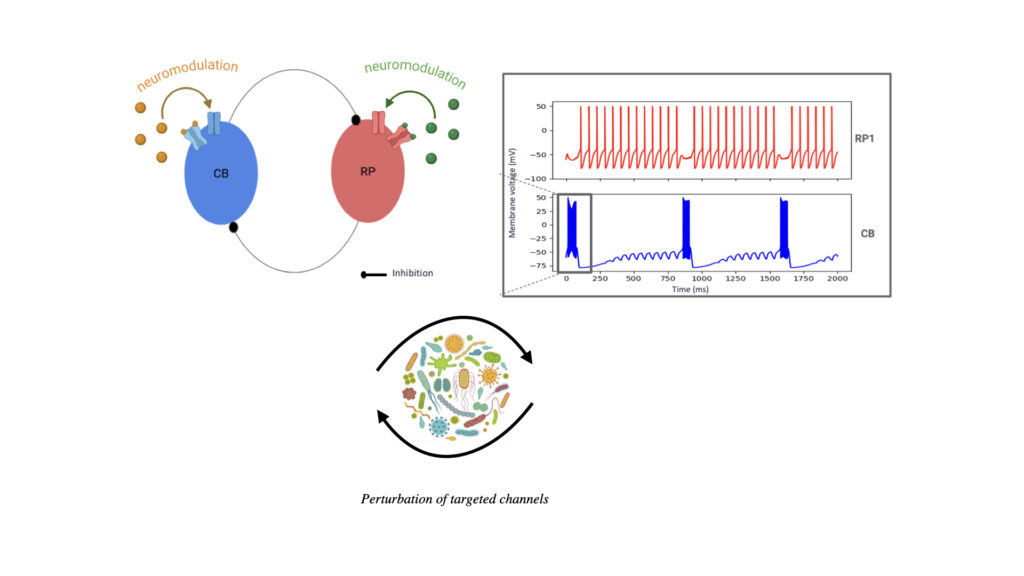
With 200–2,000 neurons spanning 11 cell types, Hydra exhibits diverse quantifiable behaviors such as contractions, locomotion, feeding, and learning. Its rhythmic neural activity makes it a valuable model for studying central pattern generators, which are also relevant to human physiology: recent studies have shown that disruptions of the gut microbiota can impair pacemaker rhythmicity and contribute to abnormal peristalsis. Hydra’s transparency and regenerative capacity make it uniquely suited for long-term imaging and perturbation studies. Using genetically engineered GCaMP Hydra, we aim to monitor neural activity and investigate its modulation by molecular factors such as neuropeptides and the Hydra microbiome.
Host–pathogen interactions at the earliest stages of infection.
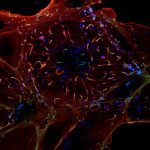
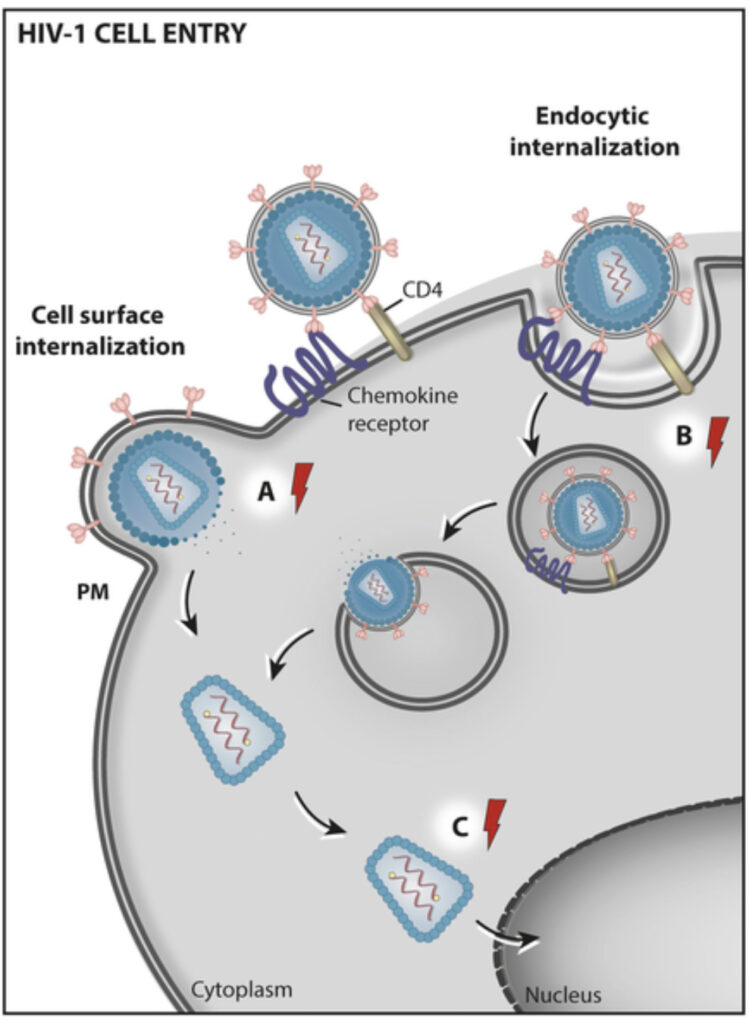
We focus on how viruses such as SARS-CoV-2 and HIV hijack cellular machinery, using advanced live imaging to study receptor and viral dynamics in space and time. These studies are carried out in collaboration with experimental partners specialized in experimental infection biology (e.g. Dynamics of Host–Pathogen Interactions Unit).
Methodological Axes
1- Mathematical modeling of dynamic networks
Using first-principles and stochastic analyses, we study how stable biological processes emerge from local interactions and how microbes perturb them. In Hydra, we build predictive neuronal models, from minimal oscillators to multi-neuron simulations derived from scRNA-seq data, to analyze how rhythms are modulated by neuropeptides, microbes, and environmental inputs. In parallel, we reconstruct the temporal choreography of viral entry by tracking receptors and viral particles and modeling key biophysical determinants.
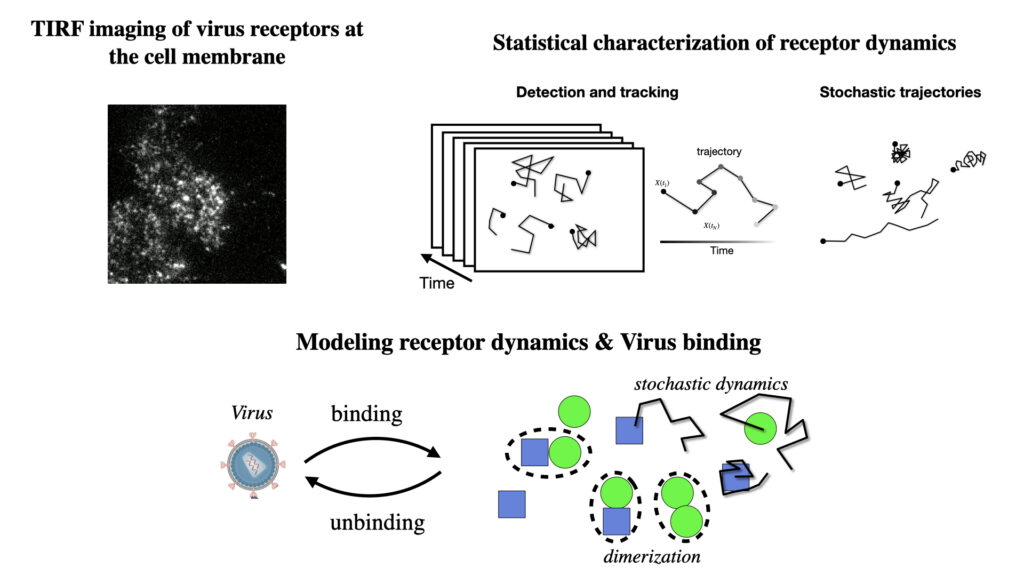
2- Statistical analysis of spatial and temporal relations between agents
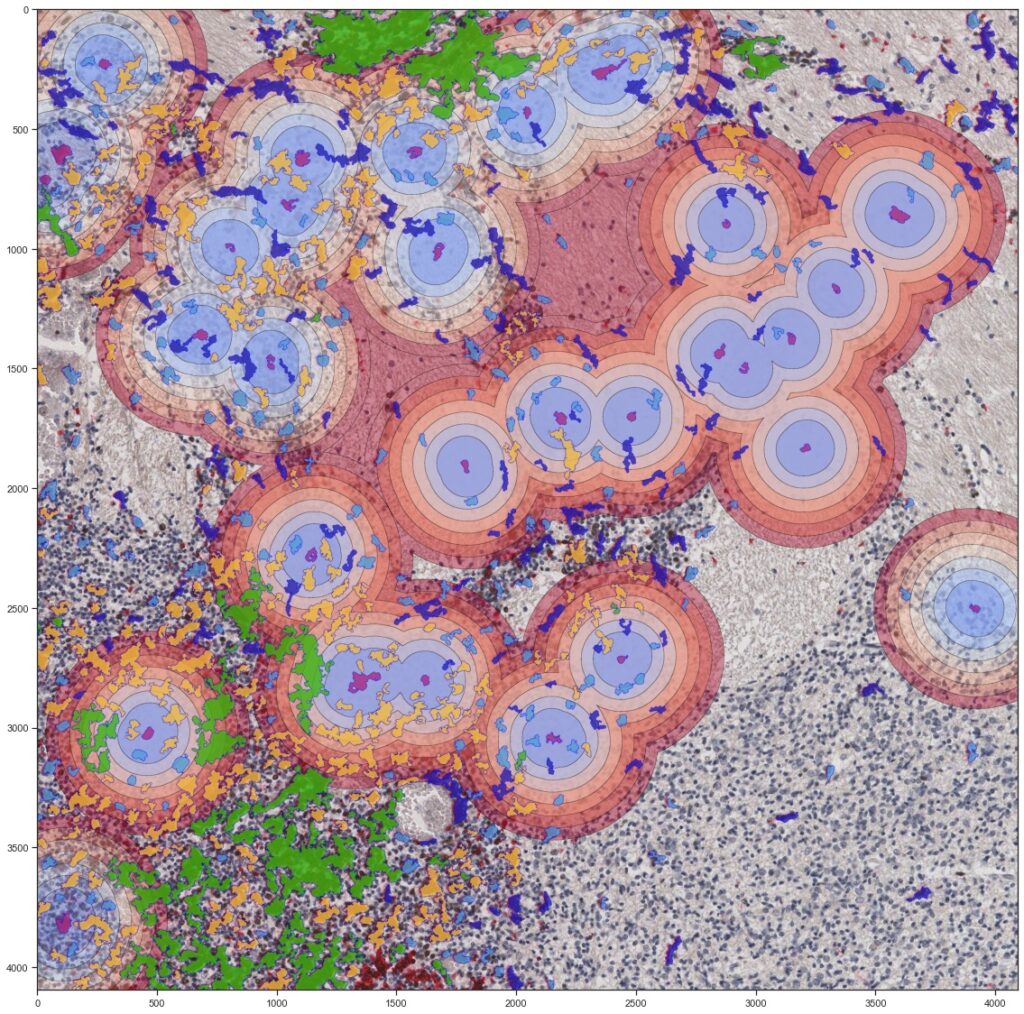
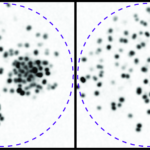
We identify mechanistic patterns in complex datasets by characterizing neuronal ensembles in Hydra and mapping host–pathogen interactions, where receptors, viral particles, and signaling complexes must be coordinated in space and time. Building on our point process framework, we generate multiscale maps linking neuronal firing to behavior and molecular interactions to cellular homeostasis or viral hijacking.
3- AI-assisted image quantification
We develop next-generation algorithms to extract quantitative information from imaged processes. In particular, we design tracking algorithms to follow thousands of objects – neurons in Hydra or single virions (or bacteria) and receptors at membranes. Our framework combine AI-based recognition with probabilistic filtering for robust performance under tissue deformation and crowding.