This is a joint team between the Institute of Biology of the Ecole Normale Supérieure and Institut Pasteur.
Emerging and re-emerging epidemics are an increasing public health threat. Climate change and human movements help spread infectious diseases. Understanding the drivers of viral and bacterial pathogenicity, their evolution and spread is an urgent task yet to be completed.
Due to recent advances in genome sequencing, millions of pathogen genomes are now available. A plethora of metadata (sampling locations, treatment history, etc.) is generated at the same time. We develop computational methods that integrate meta- and sequence data to infer spatio-temporal history and drivers of pathogen spreads, identify current epidemic trends (e.g. drug-resistant clusters) and estimate epidemiological parameters (e.g. Re), which can inform health policies. In collaboration with virologists and epidemiologists, we apply these methods to study spread of HIV, Zika, Cholera, SARS-CoV-2, Rabies and other pathogens.
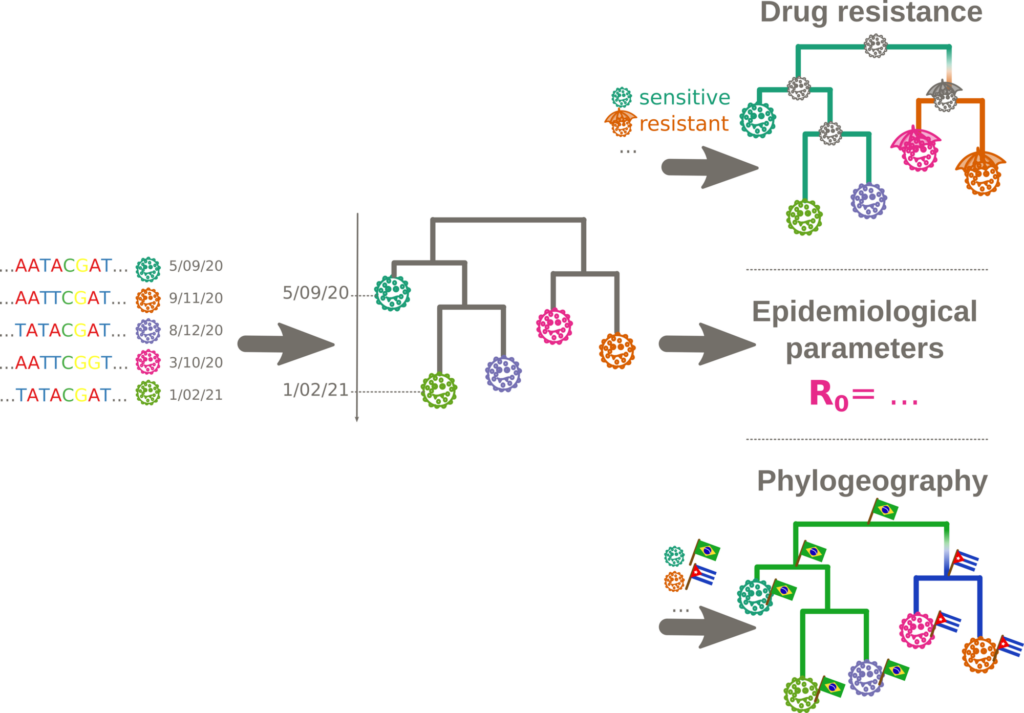
Main research directions: Our aim is to develop a methodological framework, combining metabolic modeling and phylogenetics (phylodynamics, phylogeography), applicable to large pathogen sequence datasets, and leveraging metadata to inform the analyses. It will be built on 3 main axes: (1) reconstruction of phylogenetic trees for large heterogeneous pathogen genomic datasets; (2) estimation of epidemiological parameters and spatio-temporal history of pathogen spreads from phylogenetic trees; (3) reconstruction of unknown and ancestral microbial metabolic network models using species’ evolutionary relationships in phylogenetic trees.