Nuclear Magnetic Resonance (NMR) and Hydrogen/Deuterium eXchange followed by Mass Spectrometry (HDX-MS) are powerful and versatile techniques to tackle challenging biological questions and offer complementary information to X-ray and Electron Microscopy.
Nuclear Magnetic Resonance (NMR) and Hydrogen/Deuterium eXchange followed by Mass Spectrometry (HDX-MS) are powerful and versatile techniques to tackle challenging biological questions and offer complementary information to X-ray and Electron Microscopy.
The Nuclear Magnetic Resonance (NMR) activity at Institut Pasteur
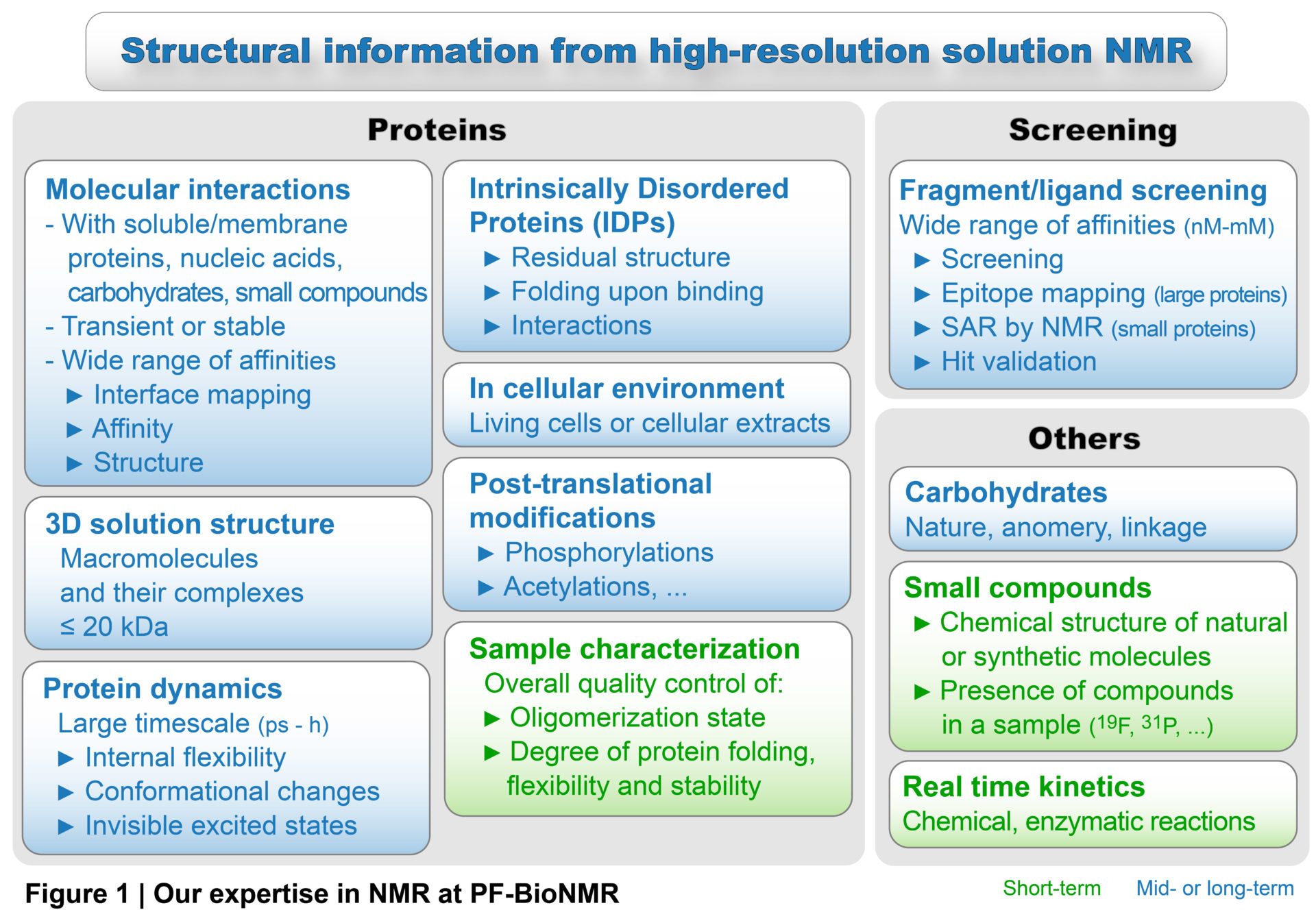
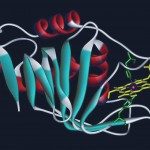
PF-BioNMR provides with state of the art NMR high-field spectrometers and knowledge to study biological macromolecules, their structure, dynamics and interactions at atomic resolution. NMR (equipment and expertise) can be used for fragment or drug-screening and drug-design, to characterise intrinsically disordered proteins or polysaccharides, to follow post-translational modifications in cell extracts, to monitor reactions in real-time or to determine the chemical structure of natural or synthetic molecules (Figure 1).
For more information on the NMR activity, you can contact us at: bionmr@pasteur.fr
The Hydrogen/Deuterium eXchange Mass Spectrometry (HDX-MS) activity at Institut Pasteur
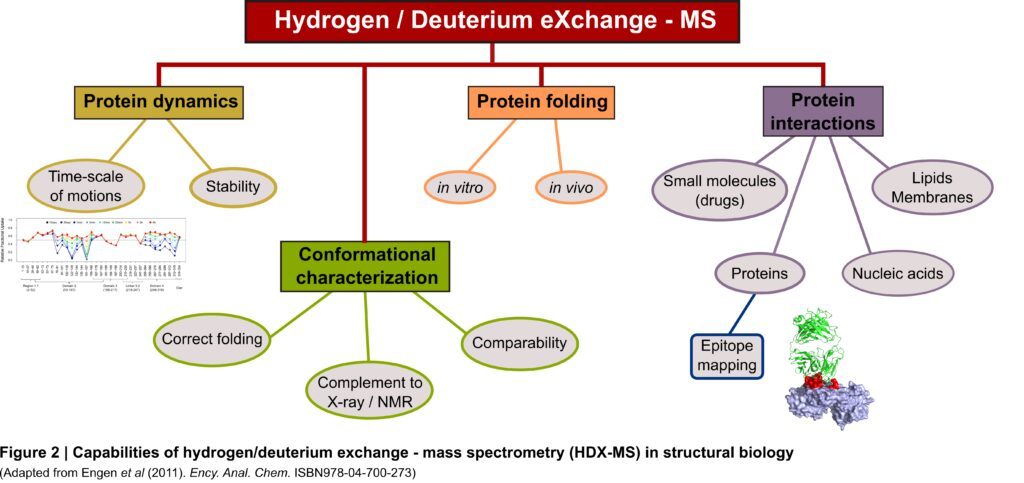
Hydrogen/Deuterium eXchange followed by Mass Spectrometry (HDX-MS) is a powerful and recognized biophysical tool in structural biology capable of probing protein/ligand interactions, conformational changes, and protein folding and dynamics (Figure 2). HDX-MS measures the changes in mass imposed by the replacement of backbone amide hydrogens by deuterium added in excess in the surrounding environment. The rate of exchange relies on the folding and dynamics of proteins making backbone amide hydrogens excellent structural probes (more details about the principle of hydrogen exchange can be found here).
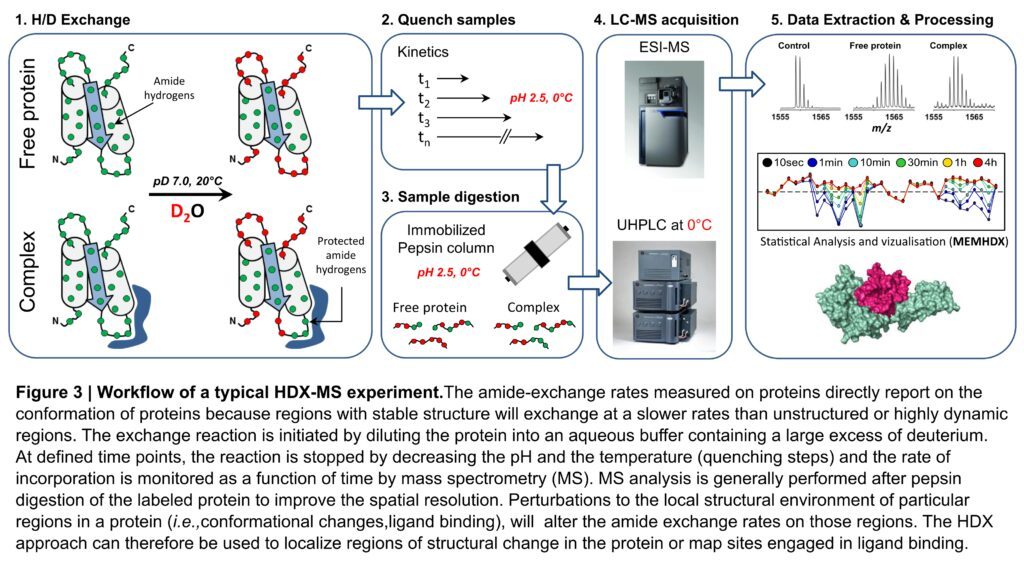
We are currently using an optimized version of the HDX-MS system commercialized by the Waters Company composed of a Synapt G2-Si HDMS mass spectrometer with ETD and IMS capabilities, a LEAP-Pal robot for automated sample handling, a cooled HDX manager that maintains the valves, columns and associated tubings at 0°C and Class M nanoACQUITY UPLC pumps (Fundings: CACSICE Equipex, C2RT Institut Pasteur). Our pipeline has been completed with the development of a software named “MEMHDX” to aid in the rapid statistical validation and visualization of large HDX-MS datasets. A schematic overview of a classical HDX-MS experiment performed with our optimized HDX-MS workflow is presented in Figure 3.
The majority of our HDX-MS projects focus on soluble, membrane-associated or integral membrane proteins that are either involved in infectious disease or considered as potential drug targets. Most of these projects are carried out in collaboration with internal or external collaborators. Our expertise in HDX-MS include:
- Analysis of the conformation and dynamic of intrinsically disordered proteins (IDPs) and large proteins (> 150 kDa).
- Analysis of small molecule/protein interactions.
- Analysis of protein/protein interaction (e.g., activation of the CyaA toxin) and identification of binding sites (e.g. Epitope mapping for both the academic and pharmaceutical sector).
- Analysis of protein/DNA interactions.
- Analysis of the conformation and dynamic of integral membrane proteins (e.g., the human glutamate transporter EAAT1).
- Optimization and improvement of the HDX-MS pipeline (e.g. MEMHDX; details about the principle of the software can be found here).
For more information on the HDX-MS activity, you can contact us at: bionmr@pasteur.fr
Click here to obtain the C2RT booklet
Click here to obtain the map of the C2RT technological core facilities