About
The genus Leptospira includes >60 Leptospira species and more than 300 serovars have been described. Little is known about the circulating etiological agents of leptospirosis in most of the regions of the world. Detailed characterization of Leptospira isolates is important for understanding the epidemiology of leptospirosis (Leptospira serovars are usually associated with a restricted number of animal reservoirs). Local Leptospira isolates can serve as antigens for the serodiagnosis of leptospirosis. The diverse distributions of Leptospira serovars and genotypes may have implications for vaccine design and efficacy.
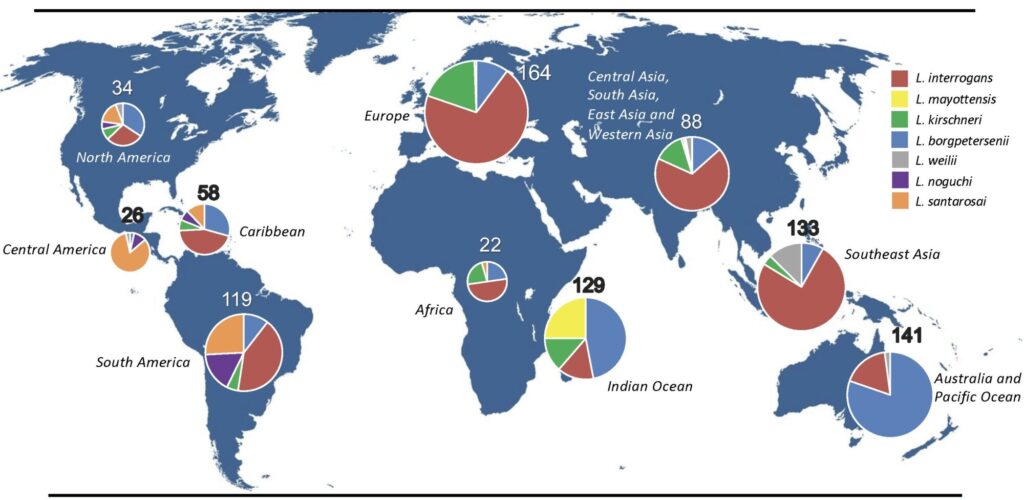
Fig. Geographic origins of the most frequent pathogenic Leptospira species in our genome database (n=917). Each pie chart corresponds to a given world region. As shown in our map, L. santarosai (n=64) is mostly found in America (North America, Central America, South America and the Caribbean islands).
To advance our understanding of the biodiversity of Leptospira strains at the global level, we evaluated the performance of whole-genome sequencing (WGS) as a new genotyping tool. We propose a set of 545 highly conserved loci as a core genome MLST (cgMLST) genotyping scheme targeting the entire Leptospira genus, including non-pathogenic strains. Phylogenetic analysis showed that cgMLST defines species, clades, subclades, clonal groups and strains, with high precision. New Leptospira species were identified from the environment. The cgMLST scheme also enables the efficient identification of serogroups and closely related serovars. cgMLST analysis demonstrated congruence with current MLST schemes, but with much improved resolution and with wider applicability, as non-pathogenic lineages were not typeable by classical MLST. In conclusion, the proposed cgMLST scheme allows high-resolution genotyping of Leptospira isolates across the phylogenetic breadth of the genus. The unified nomenclature of cgMLST genotypes, available publicly at http://bigsdb.pasteur.fr/leptospira, should enable global harmonization of Leptospira genotyping, strain emergence follow-up and novel collaborative studies of the epidemiology and evolution of this emerging pathogen.