About
The evolutionary roles of repeats
Stress is often faced in nature, whether from lack of nutrients, arrival of competitors or predators, or the presence of toxic substances. The survival of stressed organisms can be facilitated by the induction of a stress response that is specific to the individual stressor (or the class of stress encountered), by a general stress response or by generating variability allowing the recovery of an adaptive change. Dedicated stress responses are stably kept only if the stresses they tackle are sufficiently frequent. Not all stresses can be tackled in a deterministic way. The reaction to the stress must sometime include a stochastic element allowing the generation of a range of responses. In such cases, adaptation depends on the development of strategies aimed at generating appropriate stochastic variability required for natural selection. Much of this variability is generated by intra-chromosomal recombination between repeated DNA sequences. This is why I’ve been studying the presence and evolution of repeats in bacterial genomes. Recombination between repeats has been shown to play a major role in the host-parasite association between humans and bacterial pathogens. Recombination in the genomes of the latter allows the variation of proteins that are targeted by the immune system, and to exploit different host polymorphisms and tissues. Interestingly, the immune system also uses recombination to generate variability allowing it to counteract the action of pathogens. Thus, the arms race between bacteria and the immune system leads to stress in both organisms, which is partly tackled in a similar way, by stimulating recombination capable of generating adaptive changes. Our work has focused on census of repeats in genomes, on the role of repeats in sequence variation in pathogens and in the role of repeats in generating mutator genotypes.
Repeats census in prokaryotic genomes
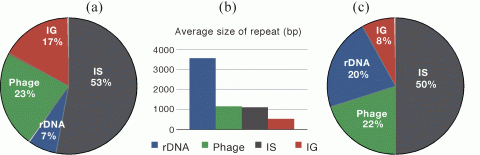
There are many repeats large enough to engage in homologous recombination in most (but not all) bacterial genomes (Figure 1). We have searched for these in over 700 genomes of prokaryotes with a set of tools that were recently put together in the programs Repseek and Repeatoire. These analyses have revealed the immense diversity of repeats in genomes, from those created by selfish elements to the ones used for protection against selfish elements, from those arising from transient gene amplifications to the ones leading to stable duplications. Experimental works have shown that some repeats do not carry any adaptive value, while others allow functional diversification and increased expression. All repeats carry some potential to disorganize and destabilize genomes. Since recombination and selection for repeats vary between genomes, the number and types of repeats are also quite diverse and in line with ecological variables, such as host-dependent associations or population sizes, and with genetic variables, such as the recombination machinery. From an evolutionary point of view repeats represent both opportunities and problems. We have therefore described how repeats are created and how they can be found in genomes.
Intragenic repeats and protein quaternary structures
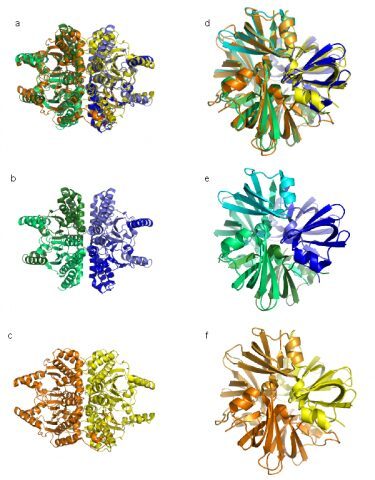
The biologically active state of many proteins requires their prior homo-oligomerisation. Such complexes are typically symmetric, a feature which has been proposed to increase their stability and facilitate the evolution of allosteric regulation. We wished to examine the possibility that similar structures and properties could arise from genetic amplifications leading to internal symmetric repeats. For this we identified internal structural repeats in a non-redundant PDB subset using Swelfe. While testing if repeats in proteins tend to be symmetric, we find that around half of the large internal repeats are symmetric, most frequently around a rotation axis of 180°. These repeats were most likely created by genetic amplification processes because they show significant sequence similarity. Symmetric repeats tend to have a fixed number of copies corresponding to their rotational symmetry order, i.e. 2 for 180° rotation axis, whereas asymmetric repeats are in longer proteins and show copy-number variability. When possible, we confirmed that proteins with symmetric repeats folding as an n-mer have homologs lacking the repeat with a higher oligomerisation number corresponding to the rotation symmetry order of the repeat. Phylogenetic analyses of these protein families suggest that typically, but not always, symmetric repeats arise in one single event from proteins that are homo-oligomers. These results suggest that oligomerisation and amplification of internal sequences can interplay in evolutionary terms because they result in functional analogues when the latter exhibit rotational symmetry.
Evolution of transposable elements
Insertion sequences (ISs) are the smallest and most frequent transposable elements in prokaryotes where they play an important evolutionary role by promoting gene inactivation and chromosome rearrangements. Their genomic abundance varies by several orders of magnitude for reasons largely unknown and widely speculated. We thus used genome data to test many of the previously proposed hypotheses, notably that IS abundance correlates with the frequency of horizontal gene transfer, genome size, pathogenicity, non-obligatory ecological associations and human-association. We re-annotated ISs in 262 prokaryotic genomes and tested these hypotheses showing that when using appropriate controls, there is no empirical basis for IS-family specificity, pathogenicity or human-association to influence IS abundance or density. Horizontal gene transfer seems necessary for the presence of ISs, but cannot alone explain the absence of ISs in more than 20% of the organisms, some of which showing high rates of horizontal gene transfer. Gene transfer is also not a significant determinant of the abundance of IS elements in genomes, suggesting that IS abundance is controlled at the level of transposition and ensuing natural selection and not at the level of infection. Prokaryotes engaging in obligatory associations have fewer ISs when controlled for genome size, but this may be caused by some being sexually isolated. Surprisingly, genome size is the only significant predictor of IS numbers and density. Alone, it explains over 40% of the variance of IS abundance. Since we find that genome size and IS abundance correlate negatively with minimal doubling times we conclude that selection for rapid replication cannot account for the few ISs found in small genomes. Instead, we show evidence that IS numbers are controlled by the frequency of highly deleterious insertion targets. Indeed, IS abundance increases quickly with genome size, which is the exact inverse trend found for the density of genes under strong selection such as essential genes. Hence, for ISs, the bigger the genome the better.



