About
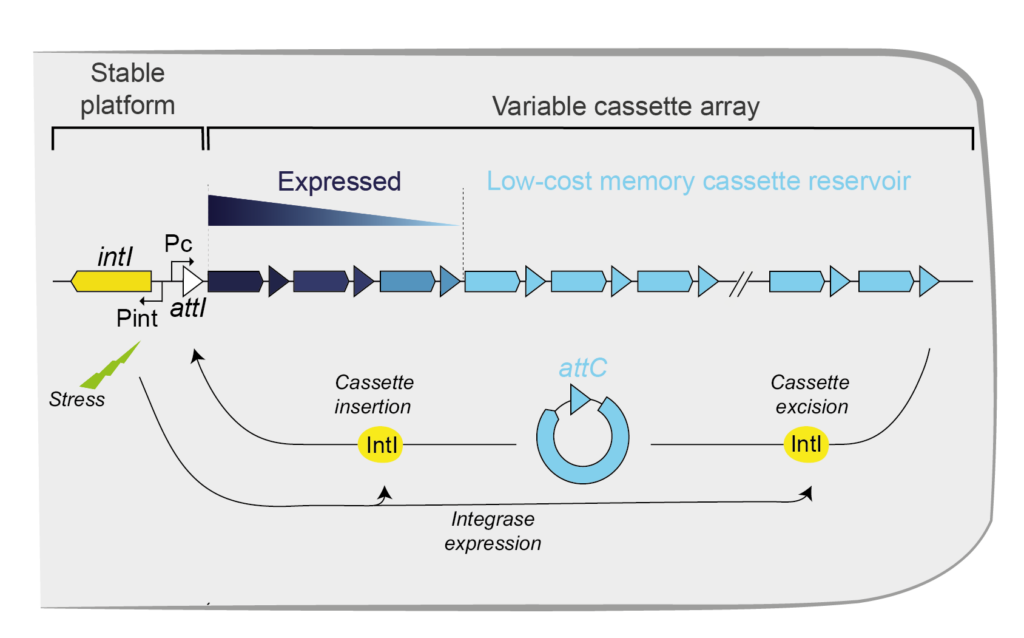
Integrase gene (intI), integrase protein (IntI), cassettes (colored arrows), promoter of cassettes (Pc) and, attachment sites of the cassettes (attC) and of the integron (attI) are shown.
Bacteria face many unexpected challenges, which require responses that may be difficult to prepare. This is often the case of antagonistic biotic interactions, e.g. when phages infect bacteria, which are regulated by evolutionary processes favoring genetic diversification. To cope with such challenges, bacteria have evolved a large diversity of mechanisms, e.g. anti-phage defense systems. These systems are extensively exchanged within bacterial communities by horizontal transfer presumably because they evolve under balancing selection, i.e. they are adaptive in certain environmental conditions, in certain moments in time, or at a given frequency in the population, but costly otherwise. As a result, while their acquisitions can be favored under certain circumstances, these systems can also be rapidly lost when their costs are not sufficiently compensated by the defense they provide. Yet, some bacteria encode a genetic system that provides a natural reservoir for these functions: the integron . Integrons are composed of a specific integrase (IntI) and an array of gene cassettes flanked by recombination sites (attC) (Figure 1). Usually, cassettes are devoid of promoters and are therefore expressed only when located at the beginning of the array thanks to the Pc promoter. Under stress conditions, the integron integrase is expressed leading to the capture of new cassettes and to rampant cassette excision and re-integration at the beginning of the array (in the attI site) (3). This mechanism allows to sample the combinatorial phenotypic diversity encoded in the integron when it may of highest value (when bacteria are under challenging conditions). Novel adaptive configurations of the cassette array may then increase in frequency in the population by natural selection. This process facilitates adaptation by circumventing time-consuming de novo evolutionary innovation and the hazardous process of acquiring novel genes by horizontal transfer (Figure 1). Some integrons can be carried by conjugative plasmids making them highly mobile. These so-called Mobile Integrons (MIs) have disseminated in many environmental niches and are now widespread in hospitals and community worldwide. MIs have been found associated to 190 antimicrobial resistance (AMR) cassettes comprising resistance determinants against almost all antibiotic families. MIs are relatively small since they contain no more than eight cassettes. Much larger sedentary and chromosomal integrons (SCIs) are present in the genomes of about 10% of sequenced bacteria. Some SCIs constitute a huge cassette reservoir and may contain up to 300 cassettes. If exchanges between SCI and MIs are frequent, they could contribute to the remarkable adaptive success of the latter. Yet, twenty years after their discovery, there are still huge knowledge gaps on the functions of the large repertoire of genes encoded by SCIs, their rates of shuffling within the integron array, their rates of transfer to MIs, and how these functions can be stabilized in genomes. These gaps prompted the Definbank research program, which combines experimental and computational approaches to elucidate the role played by SCIs in bacterial adaptation.