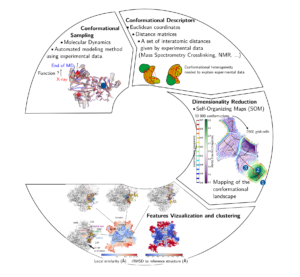
Proteins are large macromolecules playing a variety of functions within the cell. The function of the protein is linked to its three-dimensional structure. However, a protein is not a static object and the structural dynamics of a protein plays a large role for the function. Conceptually, the different conformations a protein adopts can be synthesized in term of energy landscape. Highly populated states can be seen as deep energy wells and the kinetics of transitions between them are related to the high of the barriers between the wells. However, the high dimension of the protein conformational space impairs the representation and the analysis of that landscape.
I develop dimensionality reduction techniques to unravel protein structural motions and link the dynamics to the function played by a protein. I use a combination of pure theoretical approaches based on molecular dynamics (MD) simulations and other sampling approach driven by experimental data. Among other approaches, I develop the mapping of the protein conformational space based on Self-Organizing Maps (SOM).