About
This project aims at understanding the synaptic environment at the single receptor scale. Now that large amounts of dynamical single molecule data are available and that single molecule science is slowly transitioning into a big data science, new statistical methods can be developed to investigate cellular environments at the nanometer scale. We combine random walk theory, Bayesian inference and machine learning to simultaneously probe the properties of the synaptic environment and the features of receptor dynamics. This project builds on the InferenceMAP approach.
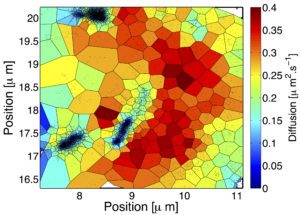
Here is an example diffusion map of Glycine receptors from data recorded by Charlotte Salvatico and Christian Specht from Triller’s lab at ENS.
We address several challenges to optimize the inference.
-
- Inference without tracking for high particle densities.
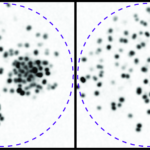
Modern time-resolved super-resolution localization microscopy techniques, e.g., PALM and uPAINT, make it possible to record the dynamics of single biomolecules at the whole-cell level at fast video rates (~100 Hz) and high tracer densities (1-10 μm-2). This in principle provides enough statistics to resolve both the spatial heterogeneity and the time-evolution of intracellular dynamics. However, a crucial step in all traditional analysis pipelines – the assignment of molecule trajectories (tracking) – breaks down at high tracer densities (MPA in the figure below). We leverage graphical models and belief propagation to assign trajectories probabilistically and remove the need for tracking, enabling accurate inference of the underlying dynamics where tracking fails (BP in the figure below). - Bayesian inference and the Ito-Stratonovich Dilemma.
The typical inverse problem in single-molecule dynamics consists in inferring the parameters of the environment (such as diffusivity and local forces) from the observed trajectory within a framework of a chosen model. One of the popular inference models is a random walk, generated by the Overdamped Langevin Equation, which ignores the inertia of the moving particle. Although this model provides an easy way to perform calculations, it comes with a drawback that the stochastic integral appearing in the Overdamped Langevin Equation is ambiguously defined. Specifically, if the interpretation of the stochastic integral used in the inference method (the inference model) is different from the one used to generate the trajectory of the particle (the generative model), the inferred map of local forces may be shifted from their real values by some bias. This effect is known as spurious forces.This problem does not appear when the integral interpretation used to generate the trajectory is known, or when the diffusivity of the particle is uniform. However, both of these assumptions likely fail in real biological systems. So when analyzing real experimental data, one has to question whether the inferred local forces are due to active force-generating processes or simply due to a spatial variation of the diffusivity. Our group is working on a method that will allow one to calculate the full posterior probability that the observed force maps corresponds to active physical forces. - “Pipeline” approaches to synapse analysis.
We are currently developing “pipeline” approaches to handle large amount of data and to entirely automate data analysis. Proofs of concept of the pipeline analysis for synapses can be found here. Proofs of concept of the pipeline analysis for time evolving processes, namely the GAG dynamics during the Virion formation, can be found here.Below is shown an example of the results of the pipeline analysis for synapses. The figure shows cumulative distributions of trapping energies (A) and diffusivities (B) of wild type (WT) receptors (black) and WT– (without beta-loop, in orange) from several hundreds of synapses.

- Finally, we are developing a software platform, TRamWAy (under heavy development) to entirely automate analysis of single molecule experiments (Coming Soon …)
- Inference without tracking for high particle densities.