Présentation
Membres

Publications
Télécharger-
2024Structural basis of TMPRSS2 zymogen activation and recognition by the HKU1 seasonal coronavirus., Cell 2024 Jun; (): .
-
2024Cryo-EM structures of type IV pili complexed with nanobodies reveal immune escape mechanisms, Nature Communications, 2024, 15 (1), pp.2414. ⟨10.1038/s41467-024-46677-y⟩.
-
2023TMPRSS2 is a functional receptor for human coronavirus HKU1., Nature 2023 Oct; (): .
-
2023Anti-V1/V3-glycan broadly HIV-1 neutralizing antibodies in a post-treatment controller., Cell Host Microbe 2023 Jul; (): .
-
2022Tunneling nanotubes provide a route for SARS-CoV-2 spreading., Sci Adv 2022 Jul; 8(29): eabo0171.
-
2022Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome, Cell 2022 Apr; 185(8): 1297-1307.e11.
-
2022Atomic structure of Lanreotide nanotubes revealed by cryo-EM., Proc Natl Acad Sci .
-
+Voir la liste complète de publications
How to Submit
For submissions, please read the below section carefully, read the full application form carefully and fill it in. Send the filled in form to: nanoimaging@pasteur.fr
It has been decided that for the initial phase of startup we will be accepting projects on a “first-come-first-served” basis, in order to maximize the data collection and allow full flexibility. This means that for the time being there will be no selection of projects. All projects will be considered to be of equal importance and all projects will be allowed microscope time. These projects can be submitted independently or in collaboration with Dorit HANEIN. The order at which these projects are imaged will depend on the date of their submission. We will also introduce a fair use policy, meaning that a new project from every unit can only be submitted after the previous project has been imaged. In other words: every unit will be allowed one submission. The second submission can be entered only after the first has been imaged. A unit is defined as researchers reporting into the same unit PI. If a unit has multiple group that have projects that require cryo-EM, then it is up to the Unit head to decide priority within the unit. Initially, we will queue all projects and assign imaging days as they become available. Once we have setup a reliable workflow we will open a PPMS agenda for users to select and book specific days. We will closely monitor the number of submissions and will evaluate if there is an overload of projects and a need to start introducing a selection committee for project prioritization or change the way we operate. In the initial startup phase we will rely on some flexibility until we can settle on a routine mode of operation. Please be aware that many projects require screening of grids first and it could be that all screened grids will not be good enough for data collection! However, a unit will have a full day on the microscope. It is therefore advised that if a unit has multiple PI’s/group leaders which all have projects for EM, they will have these different samples lined up. If it turns out that the first project does not have optimal samples yet, a second project of the same unit can be imaged. It should also be mentioned that the Titan can have a very high throughput for especially SPA projects, given the right type of sample. In some cases a full data set can be acquired within a few hours. Also in this case we will be able to run several projects form one unit in a single 24h session. For more guidance and advice please come to the NanoImaging core so we can discuss the best strategy for most effective data collection. During the first months of operation we will have trained staff at your disposal that can help prepare and image your samples. However, for future operation we will encourage each user to be trained to use the facility independently and we will be charging extra fee for an operator. We project to have frequent training sessions both on sample preparation and on microscopy imaging. In preparation of these training sessions and in order to make them as effective as possible, potential users will need to register and take an online course (www.EM-Learning.com) in order to have sufficient starting knowledge to be trained on these tools. In return, we can guarantee that by the end of the online course followed by a hands-on-course at the NanoImaging Core, users will be fully independent operators in cryo-EM. Consumables needed for hands-on training need to be ordered by each group independently. The core will only have spare consumables available. A list of consumables will be sent along with this communication. All samples that will be imaged at the core facility must be BSL1! The project submission form can be downloaded here The user order list can be downloaded here
Equipment
The NanoImaging Core Facility equipment is divided into two different categories: Sample preparation and Imaging. In this section you will find updates on the current status and performance of the different tools, scheduled and unscheduled downtime and a proof of meeting specifications. It is important to realize that the quality of final results obtained from cryo-EM will depend predominantly on the quality of the specimen. All the microscopes installed at the core facility are specified and performing well beyond the resolution that is currently achieved routinely in LS cryo-EM applications. The Titan Krios is specified to achieve a minimum of 1.4 Å resolution, while routinely showing 1.2 Å in the information limit test. To date most structures reported in the electron microscopy data bank (EMDB) are still above 5 Å resolution (~65%), and with the remaining 35% being between 3-5 Å resolution. A minority of the samples is so stable and optimized in sample preparation that is able to be resolved between 2-3 Å. The Glacios microscope is specified to 2.3 Å, however, also shows up to 1.4 Å optical resolution in the information limit test. Thus, all microscopes at the core facility, when measured daily to be within optical specifications, are capable of solving any structure to the desired resolution IF the sample preparation is up to par. The core is equipped with two different sample preparation tools and two different imaging tools. For SPA sample preparation we have two ThermoFisher (TF) Vitrobot’s and for CET we have a TF Aquilos cryo-Focussed Ion Beam (cryo-FIB) to prepare thin lamella from thick samples. For imaging we have two TF Glacios 200kV cryo-transmission electron microscopes (Cryo-TEM) equipped with Falcon 3 direct electron detectors and one TF 300kV Titan Krios G3 TEM equipped with a Gatan energy filter bioquantum/K3.
Sample Preparation

For SPA applications and vitrification of small samples the core is equipped with two TermoFisher Mk-IV Vitrobot’s that perform the process of vitrification of a sample also referred to as plunge-freezing. One is located in the Nocard building and one on the third floor of the Metchnikoff building. In order to use the Vitrobot it is required to watch the module on sample preparation on the EM-learning.com website and receive a subsequent hands-on training at the core facility. It should be emphasized that becoming experienced in sample vitrification requires significant practicing. Although the plunge-freeze technique can be learned relatively quickly (2 days), it will take time before users will be able to reproducible prepare high quality TEM-samples.

Imaging


Operational Status
Vitrobot I (Nocard): Operational Vitrobot II (Metchnikoff): Under installation Aquilos (cryo-FIB): To be delivered Nov 2019 Glacios I (200 kV TEM): Operational for SPA and Tomography Glacios II (200 kV TEM): To be delivered Nov 2019 Titan Krios (300 kV TEM): maintenance
Statistics (23/10/2019)
Total number of collected images by the core: 22414 Total number of collected tomograms: 0 Total number of structures produced: 2 Total number of samples screened: 88
Courses
Microscope time is limited and costly, however, to become a fully independent operator it is essential to have substantial “hands-on” experience. At the NanoImaging Core we will actively encourage researchers to get trained to be independent microscopist. In cryo-EM it is often necessary to spend significant time on a sample to select the best possible area’s for image acquisition. It is advantageous not be be limited to the working hours of the core’s operators and have more screening time available to increase the success of ones own projects. Due to an ever increasing amount of software and automation it is now possible to be trained for basic operation within a matter of days. Nevertheless, the TEM remains a complicated tool to master and understand. In order to effectively organize training courses and guarantee that each trainee will be able to independently operate the microscope after the “hands-on” training, it is essential to have a minimum basic understanding of the technology, familiarize ones selves with frequently used terminology and have a basic understanding of the microscope layout, controls and software. To this end we request new user and trainees to register and follow the online EM-instruction course: www.EM-learning.com. This course consist of roughly 70 hours of video instruction material ranging from theoretical lectures to “hands-on” demonstrations of the equipment and software that is installed at the core facility. During the “hands-on” practical training we will assume that the trainees have watched these videos in order to train them to be independent users in a matter of days.The training videos on the website will consist of two section: a “learning” section and a “workflow” section. The workflow section will show the full SPA workflow from sample preparation to data collection and will demonstrate the daily operation of a user at the NanoImaging core facility. The learning section is divided in 10 modules that will dive deeper into the individual components of the transmission electron microscope and features theoretical lectures alternated with “hands-on” microscope operation. The goal of the practical courses provided by the NanoImaging core facility will be to have user independently reproduce the daily workflow as demonstrated in the workflow section of the online course. The dates of the “hands-on” courses will be communicated in this section. We will be able to train 3 people per microscope, thus giving us the capacity of 9 people maximum per training course when fully operational.
Images & Médias
As a first test for both the Glacios I and Titan Krios, a stable test specimen was used for a full data run of the SPA workflow. It should be mentioned that the Apoferritin test sample is a very stable particle that behaves well, can be easily recognized and picked by automated software routines and in general can be reconstructed to high resolution if the sample thickness is in the order a single layer of particles (no overlapping particles in the vitreous water film). An initial run of ~10 hours on Glacios I at 200 kV equipped with a Falcon 3EC camera operating in counting mode yielded ~130.000 particles that reconstructed to 2.6Å after initial image processing and 2.4Å after additional particle polishing. After data collection the same EM-grid was transferred to the Titan Krios for further data collection using the Bio-quantum/K3, energy filter/camera combination at 300kV. Here we continue to collect for 13 hours yielding ~900.000 particles that produced a 2.4Å reconstruction before particle polishing. Due to the sheer size of the data set we submitted 1/13 part of the set for particle polishing. The 1/13 data set contained ~70.000 particles that initially reconstructed to 2.7Å resolution before particle polishing and 2.4Å after. The above shows that increasing the data set ~10 fold has about the same result as the particle polishing routine on the smaller subset of the data. It also shows that the final result obtained on the Glacios at 200kV is about the same as the data recorded on the Titan at 300kV. Both microscopes do not limit the resolution of the final reconstruction. The sample thickness and particle stability dominate the final result obtained. Although the data can be pushed further and it is expected that the full 900.000 particle data set after polishing will reach higher resolution, it should be considered that most biological questions can be answered at the resolution of 2.4A which was obtained after 1 h data collection on the Titan Krios and one data run of the Glacios equally.
Glacios I
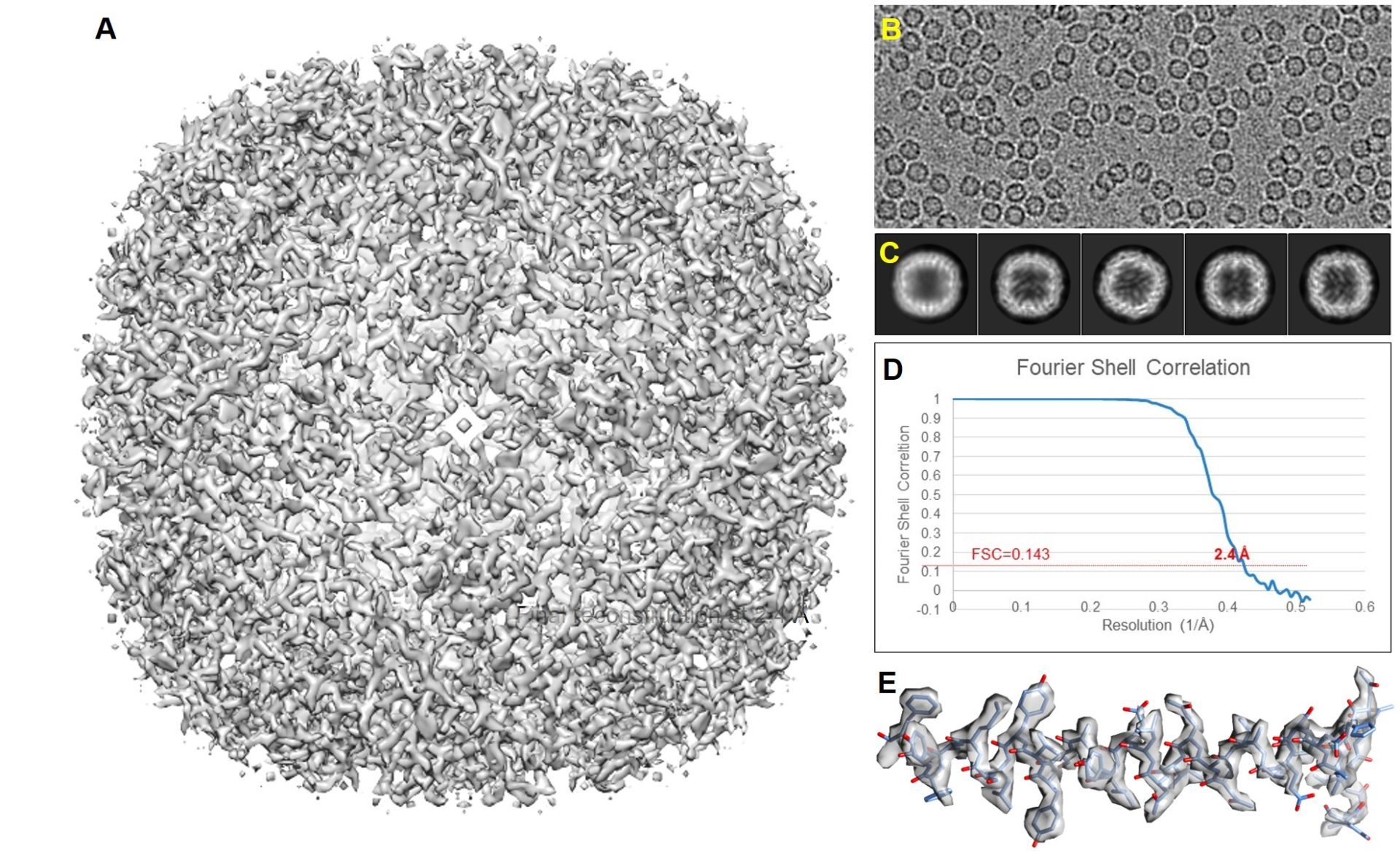
Titan Krios
First recorded test data set reconstructed to 2.4 Å SPA result on the Titan Krios. A data collection run of 13 hours yielded 2154 images in which 890.000 particles could be picked, which reconstructed to a 2.4 Å structure before particle polishing. 1/13 size of the data set (165 images) contained 68.000 particles that reconstructed to a 2.7 Å structure before particle polishing and 2.4 Å after.