Présentation
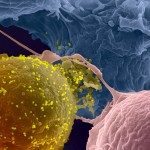
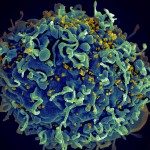
The pathogenesis of HIV infection, and in particular the development of immunodeficiency, remains incompletely understood. During HIV infection both innate and adaptive immune responses are raised, but they appear to be insufficient or too late to eliminate the virus. Whereas much knowledge exists on the role of adaptive immunity during HIV infection, it has only recently been appreciated that the innate immune response may also play an important part in HIV pathogenesis.
One can easily assume that the constant production of viral RNAs and proteins in a chronic lentiviral infection has the potential to stimulate innate immunity response and likely to over-activate the whole immune system eventually resulting in AIDS. Retinoic acid-inducible gene-I (RIG-I), melanoma differentiation factor 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2) are pattern recognition receptors that play a pivotal role in sensing nucleic acids from pathogens in the cytoplasm of infected cells and in inducing type-I interferon (IFN). Several studies suggested that RIG-I and MDA5 could detect lentiviral RNA, but currently there are no direct evidences of RIG-I or MDA5 interaction with viral RNA in HIV-infected cells and the sequences of HIV nucleic acids that are involved in these interactions are unknown.
Today new DNA sequencing technologies, such as next-generation sequencing, open a possibility of high-throughput sequencing of RNAs at low cost with no need to design specific primers. We have recently developed and validated a new protocol to characterize, in an infectious context, the nucleic acids bound to a viral protein by using a combination of virus reverse genetics, affinity purification and next-generation sequencing. In this project we wish to take advantage of this protocol to perform high-throughput identification of HIV RNA ligands for RIG-I, MDA5 or LGP2 cytoplasmic sensors.