About
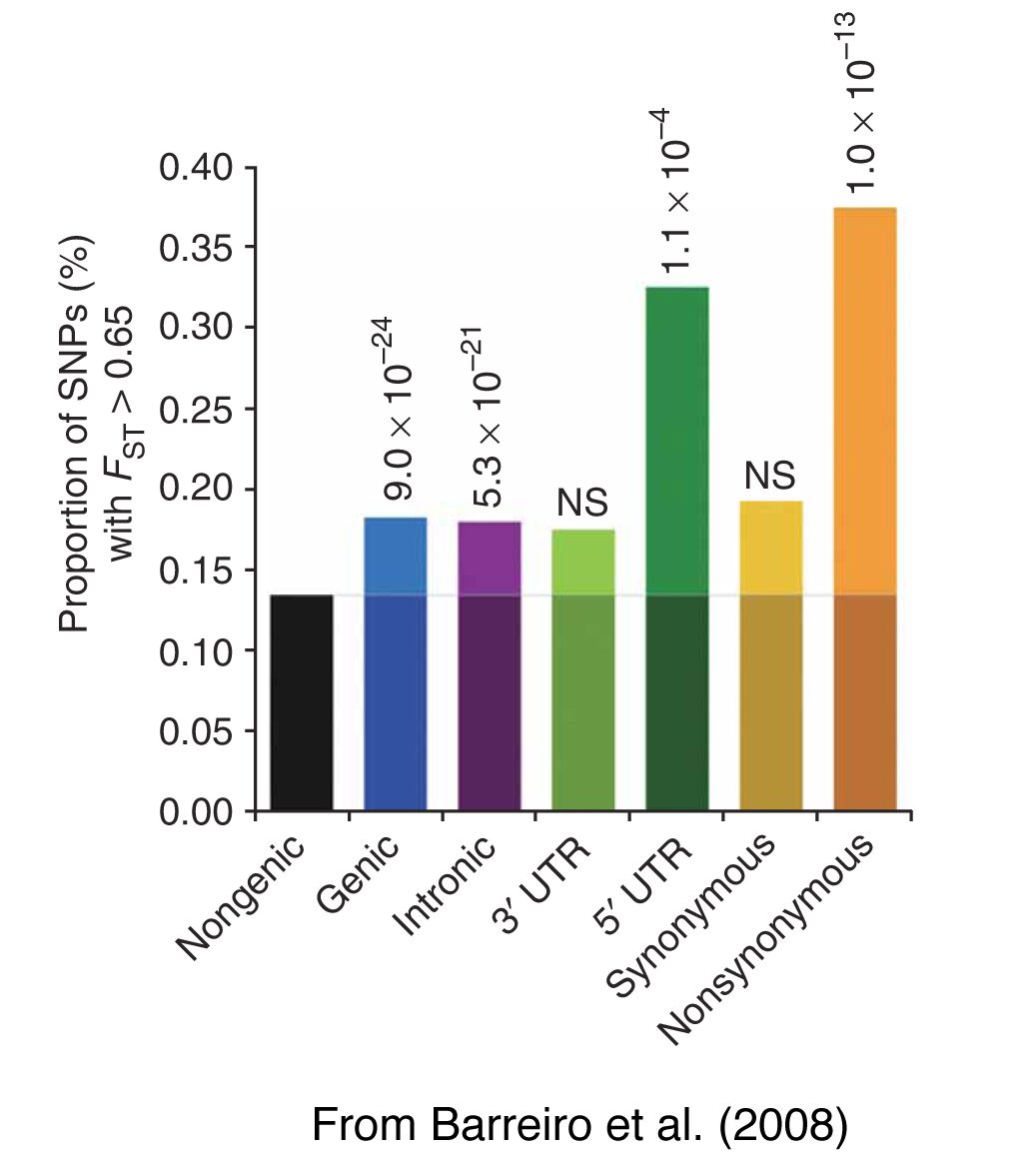
The wide range of phenotypic variation in human populations may reflect distinctive processes of selection/adaptation to variable environmental conditions (e.g. climate, pathogens, nutritional resources). The advent of genome-wide datasets allows testing different hypotheses concerning how natural selection, in its different forms and intensities, has influenced the variability of the human genome. For example, we found that negative selection has globally reduced population differentiation at non-synonymous variants at the genome-wide scale, particularly in disease-related genes. Conversely, positive selection appears to have increased population differentiation in gene regions, primarily at non-synonymous and 5’-UTR variants. Our analyses have also identified a group of genes, which show strong signatures of population-specific positive selection, having most likely participated in the processes of population adaptation to their specific environments. We are also interested in the evolutionary processes behind the patterns of variability of genomic regions involved in the regulation of gene expression, such as microRNAs. We are currently developing new statistical approaches to identify the effects of natural selection from next-generation sequencing datasets.