About
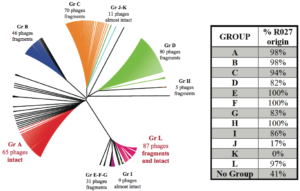
The capacity to rapidly and accurately identify virulent strains of C. difficile in order to prevent severe and fatal infections remains a major chal- lenge for clinicians and investigators. Also, the evolution of an infection and the resulting clinical issue are multifactorial, making it difficult to identify clear virulence factors or biomarkers that could help achieve this goal. Bacteriophages have previously been found to have a major influence on the pathogenic poten- tial of C. difficile, notably by their capacity to modulate the toxin production level. However, phage presence and impact on C. difficile biology is still poorly understood. Thus, we sought to make a detailed search and analysis of bacteriophage elements in the genome of 81 diverse clinical strains, mostly ribotype 027 (R027), sampled in Quebec and Ontario between 2005 and 2008. With this study, we aimed to see if phage content can be a source of genetic variability and a source of virulence bolstering elements among R027 strains, a group still widely considered as genetically homogenous.