About
The agent of epidemic dysentery, the bacterium Shigella dysenteriae type 1, was isolated in Japan in 1897 by Dr Kyoshi Shiga during a massive epidemic of bloody diarrhea that caused 20,000 deaths over a six-month period. Since then, strains have been isolated the whole world over, but without any light being shed on the origin of the epidemics caused by them or the links between them.
To provide an understanding of this disease, we have undertaken an unprecedented genomic study in collaboration with the Wellcome Trust Sanger Institute and several international institutions. Using new technologies for bacterial genome sequencing and bioinformatics, we analyzed more than 330 strains of Shigella dysenteriae type 1 isolated between 1915 – in soldiers taking part in the Dardanelles campaign – and 2011 and collected by 35 international institutes in 66 countries.
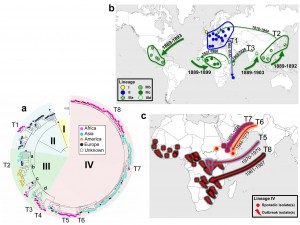
The study shows that, contrary to popular belief, the Shigella dysenteriae pathogen currently raging across Africa and Asia is most probably of European origin. The common ancestor of the most ancestral strains circulating in Europe early in the 20th century existed as far back as the 1740s, and it could have been responsible for the violent epidemics of bloody diarrhea described in Western and Northern Europe during the 18th and 19th centuries – well before the work of Louis Pasteur and studies in microbiology. These dysentery epidemics went on to kill over 200,000 people in France, mainly between 1738 and 1742 and in 1779, and 35,000 in Sweden between 1770 and 1775. In the 19th century, Ireland was also severely affected, during the great famine of 1846 to 1849, and the disease struck again in France during the Franco-Prussian War.
European expansion helped propagate the pathogen. By identifying different genetic lineages, we were able to trace the path of the bacterium worldwide over time. In well under 20 years, between 1889 and 1903, S. dysenteriae started in Europe and spread from there to America, Africa and Asia. This dissemination was probably aided by population movements during European emigration to America as well as the colonization of territories in Africa and Asia. The whole process was also likely facilitated by steam navigation and the opening of the Suez Canal in 1869.In the 20th century, the bacterium reappeared in Europe during the First World War, in particular during the Dardanelles campaign (1915-1916), where it was a significant factor in the defeat of the Allied troops. It was also identified in Central Europe during the Second World War, before dying out in Europe. However, it continued to spread across Asia, Africa and Central America in the form of violent outbreaks. The Central American epidemic of 1969 to 1972 saw 500,000 cases. The outbreak in the Indian subcontinent (India and Bangladesh) subsequently became active throughout the 20th century and would prove to be the source of several epidemic waves that spread to Africa and South-East Asia.
In the 25 years between 1965 and 1990, 99% of strains developed antibiotic resistance. Since the first bacteria were isolated well before the use of antibiotics in humans, study of the collection has also placed a precise date on the appearance of the first antibiotic resistance, in Asia and America in the mid-1960s. The bacterium then acquired resistance genes against most classes of antibiotics, and since the 1990s less than 1% of bacterial strains have remained susceptible to antibiotics. We consider it inevitable and a cause for concern that the Shiga bacillus circulating in the same regions as these resistance genes shall acquire resistance to the last-resort antibiotic classes. This pessimistic outlook may, however, be balanced against a reduction in infection since 2010, including in Southern Asia and Africa. Although the bacterium is no longer isolated during epidemic waves, it is still found during field studies, such as that carried out by Médecins Sans Frontières in diarrheal children in Niger in 2011. For Dr François-Xavier Weill, the Shiga bacillus is still circulating at a low level, and could be responsible for future epidemics if conditions should prove favorable – such as a large gathering of people without access to drinking water or treatment of human waste. He believes that this study highlights the need for an effective vaccine, which will be crucial for controlling this disease in the future in view of the reduced efficacy of antibiotics.
This study is published in the journal Nature Microbiology and is readable at http://rdcu.be/gU1g