About
Epithelial cell death is widespread during development and tissue homeostasis. While the core pathway of apoptosis is well known, we lack predictive power to understand when and where a specific cell will die in a tissue. Yet, this should have important consequences for clone selection and tissue homeostasis, as exemplified by the process of cell competition. This context-dependent cell elimination is essential for tissue homeostasis and tumour progression, but we know very little about its prevalence during normal development. Recent works have characterised multiple non-apoptotic functions of effector caspases and a high frequency of cells undergoing transient caspase activation without death. Yet, what distinguishes a dying from a surviving cell downstream of effector caspase activation is completely unknown. Using quantitative live imaging of effector caspase sensors and caspase activity tuning by optogenetics in the Drosophila pupal notum (a single layer epithelium), we found that cells engage in apoptosis at very variable levels of effector caspase activity. This suggested that the engagement in apoptosis can be developmentally modulated downstream of the most terminal caspase activation and that a completely new layer of regulation of cell death could have important physiological roles. Here, we propose to combine quantitative live imaging, multivariate model, supervised machine learning and quantitative perturbations through optogenetics to define parameters predictive of the engagement in apoptosis downstream of caspase activation. We are also using spatial omics combined with an optogenetic-based screening to identify new factors modulating the sensitivity to caspases. Eventually, we are testing the physiological functions of caspase sensitivity modulation could it be for biasing cell selection and generating spontaneous cell competition, characterising its impact on tissue morphogenesis and modulating oncogenic cell elimination.
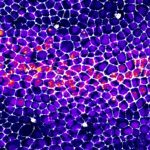
Movie of a Drosophila pupal notum showing live caspase activity (Green, GC3Ai) and cell contour (E-cad, magenta). The right channel shows the cumulative caspase activation after segmentation and image processing.