About
Organic synthesis, still the limiting factor of Medicinal Chemistry?
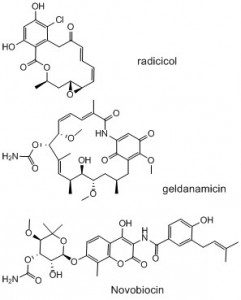
This ongoing project is based on our reviews (1-3) on the inhibitors of the ATPase functions of human heat shock protein 90 (HSP90) or bacterial type IIA topoisomerases (DNA gyrase and topoisomerase IV). Since this project has so far received only modest funding from the Institut Pasteur, and none from other agencies, the following text is more focused on strategic considerations. HSP90 and bacterial type IIA topoisomerases are fairly modern target of medicinal chemistry respectively in oncology and antibacterials, interestingly, they are sharing some common features. HSP90 is a chaperone which, via complex interactions with various co-chaperones, insures the proper folding of more than 200 cellular proteins, whereas bacterial type IIA topoisomerases are dealing with the proper compression and decompression of DNA necessary for its expression. In other words, HSP90 and bacterial type IIA topoisomerases are respectively taking care of proteins or DNA spatial organisation. Moreover, to achieve these functions, both types of enzymes have an ATPase activity which is very likely providing the energy for the extensive conformational changes that take place in the course of their action. Interestingly, the structure of the ATPase component of these enzymes are similar as they both adopt a particular fold called a GHKL or a Bergerat fold. It is probably relevant to mention that these enzymes were discovered because of naturally occurring inhibitors: radicicol and geldanamycin for HSP90 and novobiocin for DNA gyrase. Indeed, these compounds had remarkable phenotypic effects and this eventually led to the identification of their biochemical targets (drug deorphaning is not an easy job).
Of more interest is that many series of inhibitors of the ATPase functions of these enzymes have been discovered and, so far, no cross-inhibition has been reported. Moreover, the processes used to find these series are providing strong arguments for retaining medicinal chemists in any credible drug discovery program. Indeed, computer-based approaches still have some substantial progress to make to provide credible ex nihilo alternatives.
In any case, in this computer-based age, one major challenge would be to integrate the accumulated knowledge on the structures of the series of inhibitors found as well as their binding mode to the ATPase cavities. From this, a Delphic computer-based application, going beyond the current limitations and truly able to predict the binding ability/potential of a given compound would be very useful. Why this? Because, unfortunately, in far too many cases the inherent structural features of a given series of inhibitors make them not suitable for human medicine. The only options are then extensive rescaffolding strategies and the use of bioisosteric replacements to prepare related series of compounds. Knowing, with a higher degree of certainty, that these new compounds should be inhibiting the targeted enzyme would save quite a lot of time. To quote a sobering review on the problem of discovering new antibiotics: “The only way to overcome the challenges of multifactorial antibacterial lead optimization is to expand the number of chemical derivatives. We now employ roughly two chemists for each biologist in the antibacterial therapeutic area, a fourfold turn–around from the days when genomics dominated our activities” (4). Meaning: organic chemistry remains the limiting factor of drug discovery (right! back to the bench guys).
Thank you for reading
Yves L. Janin
(1) Janin, Y. L. Heat shock protein 90 inhibitors. A text book example of medicinal chemistry? J. Med. Chem. 2005, 48, 7503-7512.
(2) Janin, Y. L. ATPase inhibitors of heat shock protein 90, second season. Drug Discov. Today 2010, 15, 342-353.
(3) Mayer, C.; Janin, Y. L. Non quinolone inhibitors of bacterial type IIA topoisomerases, a feat of bioisosterism. Chem. Rev. 2014, 114, 2313-2342.
(4) Payne, D. J.; Gwynn, M. N.; Holmes, D. J.; Pompliano, D. L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nature Rev. 2007, 6, 29-40.