About
Gerald Spaeth, coordinator and PI
Eric Prina, co-PI
Manfred Jung (University of Freiburg, Germany), collaborator
Dysregulation of epigenetic control has been recognized as a major source of human disease, ranging from cancer to microbial infection. Many viral, bacterial and eukaryotic pathogens infect mammalian cells and have co-evolved strategies to modulate the expression profile of their host cells promoting their own survival [1-7]. Despite the devastating consequences for human health, the epigenetic mechanisms underlying such host cell subversion are only poorly understood. Here we apply the protozoan parasite Leishmania as a unique model system to lift this major scientific barrier by investigating the epigenetic mechanisms that allow these parasites to evade macrophage immuno-metabolic functions and to exploit these essential immune cells for intracellular survival.
This effort revealed a series of inhibitors targeting the macrophage epigenetic enzyme ‘lysine specific demethylase 1’ (LSD1, aka KDMT1a) that eliminate intracellular Leishmania in a host-directed fashion. Together these results lend strong support to our major working hypothesis that Leishmania subverts macrophage LSD1 functions to establish permissive conditions for intracellular parasite survival and that investigating LSD1 functions in our system will provide new insight into epigenetic mechanisms underlying intracellular infection and the development of pathological polarization states of macrophages.
Our findings open a series of important questions on (i) the molecular mechanisms that alter the macrophage epigenetic profile and LSD1 activity during infection, (ii) the nature of the macrophage pathways that are controlled by LSD1 and exploited by the parasite for intracellular survival, and (iii) the dynamic chromatin interactions that regulate the expression of LSD1 target genes. We will investigate our working hypotheses by synergizing the complementary expertise of the French partner in epigenetics of Leishmania/macrophage interaction and the German partner in chemical biology and the synthesis of epigenetic probes through the following objectives:
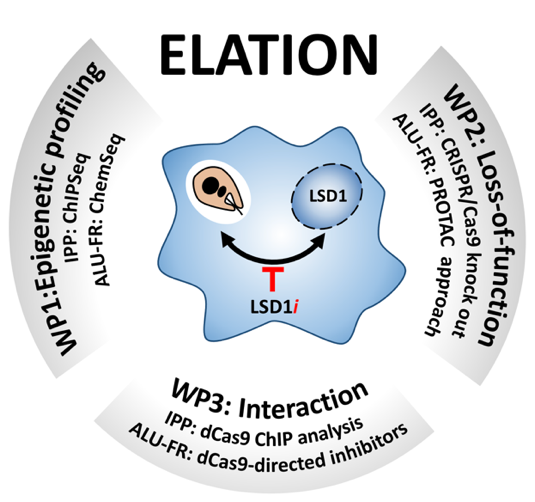
Objective 1: Mapping infection- and LSD1-dependent changes of the macrophage epigenome. We will establish a first, genome-wide map of chromatin changes in primary macrophages in response to Leishmania infection and LSD1 inhibition by (i) profiling the dynamics of chromatin accessibility using ATACseq, (ii) monitoring changes in histone H3 modification and LSD1 promoter occupancy using CUT&RUN technology, and (iii) revealing target promoters regulated by LSD1 by synthetizing and applying biotinylated LSD1 inhibitors (LSD1i) for a powerful ChemSeq approach. Integrative analyses with existing RNAseq data sets will shed important new light on regulatory mechanisms underlying subversion of the macrophage immuno-metabolomic functions by intracellular Leishmania, and restoration of the macrophage anti-leishmanial potential by LSD1i treatment. Objective 1 will further identify key macrophage promoters that are antagonistically regulated by infection and LSD1i treatment, which will be prioritized for the locus-specific LSD1 interaction analyses proposed in WP3.
Objective 2: Characterization of the role of macrophage LSD1 in intracellular Leishmania infection. We will use two complementary loss-of-function strategies (LOF) that rely (i) on the synthesis and application of LSD1i-specific chemical degraders (Proteolysis targeting chimeras – PROTACS) to eliminate LSD1 by bivalent ligand induces proteasomal degradation (ALU-FR), and (ii) the use of Cas9-transgenic, immortalized primary macrophages to generate LSD1 knock out cells (IPP). We will study the impact of chemical and genetic LSD1 ablation on Leishmania intracellular survival, on the macrophage immuno-metabolic expression profile, and on macrophage epigenetic regulation. Together these analyses will allow us to validate macrophage LSD1 as a first target for host-directed, anti-leishmanial intervention, and to further prioritize the LSD1-regulated gene promoters that will be analyzed in Objective 3.
Objective 3: Identification of LSD1-dependent, regulatory mechanisms required for parasite survival. We will (i) synthesize and apply dCas9-guided LSD1i probes that will allow for a targeted inhibition of LSD1 at regulatory DNA elements defined in Objectives 1 and 2 (ALU-FR), and (ii) apply biotinylated dCas9 for promoter-specific chromatin precipitation and the analysis of LSD1 binding partners (IPP). These analyses will allow us (i) to establish the biological relevance of individual genes and downstream-regulated pathways in LSD1i-mediated Leishmania clearance, ii) to identify qualitative and quantitative changes in LSD1 complex formation at the molecular level, and (iii) to assess the biological activity of identified complexes by correlating LSD1 interactions with the transcriptomic and epigenetic changes mapped in Objectives 1 and 2.
In conclusion, ELATION pioneers a highly innovative ChemBio approach on important open questions of intracellular microbial infection. Combining systems analysis at epigenetic and transcriptomic levels with the development of unique chemical probes will allow a first integrated view of epigenetic subversion strategies essential for intracellular Leishmania survival and will deliver a first proof-of-principle for host-directed therapy against this important pathogen. Our experimental framework will represent a useful blueprint applicable to other intracellular infections, such as tuberculosis, candidiasis or HIV and COVID.
References. (1) Pearce, E. J. and A. S. MacDonald, Nat Rev Immunol, 2002. 10.1038/nri843; (2) Herbert, D. R., et al., Immunity, 2004. 10.1016/s1074-7613(04)00107-4; (3) Noel, W., et al., Trends Parasitol, 2004. 10.1016/j.pt.2004.01.004; (4) Raes, G., et al., Curr Opin Immunol, 2007. 10.1016/j.coi.2007.05.007; (5) Benoit, M., et al., J Immunol, 2008. 10.4049/jimmunol.181.6.3733; (6) Herbein, G. and A. Varin, Retrovirology, 2010. 10.1186/1742-4690-7-33; (7) Kamhawi, S. and T. D. Serafim, Trends Parasitol, 2020. 10.1016/j.pt.2020.04.008; (8) Lecoeur, H., et al., Cell Rep, 2020. 10.1016/j.celrep.2020.01.030; (9) Marek, M., et al., PLoS Pathog, 2013. 10.1371/journal.ppat.1003645; (10) Stolfa, D. A., et al., J Mol Biol, 2014. 10.1016/j.jmb.2014.03.007; (11) Carneiro, V. C., et al., PLoS Pathog, 2014. 10.1371/journal.ppat.1004116; (12) Schiedel, M., et al., J Biomol Screen, 2015. 10.1177/1087057114555307; (13) Heimburg, T., et al., J Med Chem, 2016. 10.1021/acs.jmedchem.5b01478; (14) Monaldi, D., et al., J Med Chem, 2019. 10.1021/acs.jmedchem.9b00638; (15) Woollard, S. M. and G. D. Kanmogne, Drug Des Devel Ther, 2015. 10.2147/DDDT.S90580.
Schematic overview of the ELATION project. Our previously published and preliminary data correlate Leishmania infection with demethylation of histone H3 at pro-inflammatory promoters, causing deactivation of the macrophage immune response [8]. Our discovery of a series of anti-leishmanial compounds that target the macrophage lysine-specific demethylase 1 (LSD1, aka KDM1A) established a first functional link between Leishmania and this host epigenetic enzyme. We hypothesize that the LSD1 inhibitors (LSD1i, red bar) cause host-directed parasite killing through the restoration of an anti-microbial immuno-metabolic macrophage phenotype (see Figure 4). We will investigate this hypothesis through three interdisciplinary work packages (WPs) that synergize the complementary expertise of ELATION partners in epigenetic analysis of host/pathogen interaction (IPP, France) and chemical epigenetics (ALU-FR, Germany).