Présentation
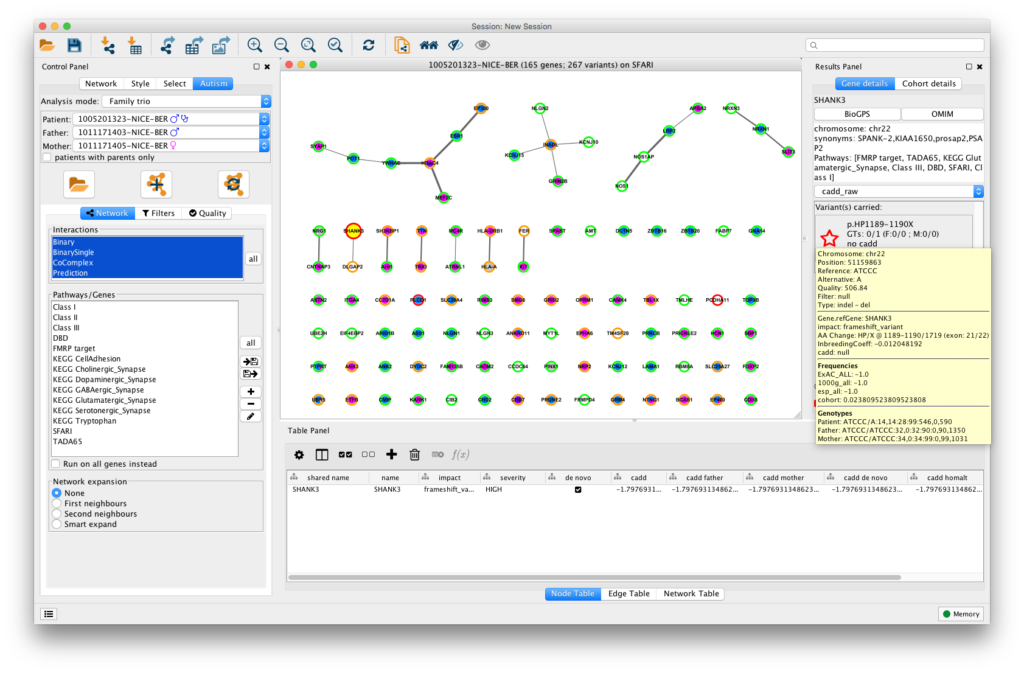
Gravity (for Gene inteRaction Analysis of Variants in Individuals: a Tool for You) provides a visualisation of genetic data from SNP array, whole genome and whole exome data in the context of protein-protein interactions. In addition, it allows the user to dynamically filter on sequencing quality or allelic frequency, or to focus on specific pathways of interest. The tool integrates analyses of mutation pattern across full cohorts of patients and their relatives, identifying multiple hits in individuals and providing a precise characterisation of the variants.
Gravity takes as an input a Gemini database generated from a variant calling file containing all the variants identified in a cohort. As an output, the visualisation of the variants found in an individual or in a family includes the network representation, the full annotation of each variant, i.e. frequency, deleteriousness and inheritance patterns, as well as cohort details on other carrying individuals. The network representation, as well as the gene annotation and sequencing parameters, can be exported through Cytoscape functions or tab-separated files.
Gravity helps in the interpretation of the combined effect of multiple mutations within different pathways of interest in the patients, setting the stage for better patient stratification and potentially more precise and personalised therapeutic strategies.
Documentation and tool available at http://gravity.pasteur.fr.