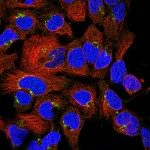
Présentation
Unraveling the structure/function of TNTs in vitro and in vivo.
While TNTs may be involved in cell-to-cell communication in many physiological processes as well as diseases, their physiological relevance is unclear as their existence in vivo has not yet been shown. Their fragility and lack of molecular markers make the observation of TNTs in vivo very difficult and has raised a lot of skepticism as to their physiological significance. Therefore, their structural and functional characterization and identification in vivo are absolutely necessary. We have demonstrated that filopodia and TNTs rely on different mechanisms of formation, different actin modifiers and Rab proteins (Delage et al., Sci Rep, 2016, Zhu et al., JCS 2018).
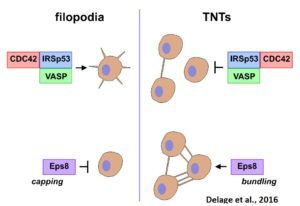
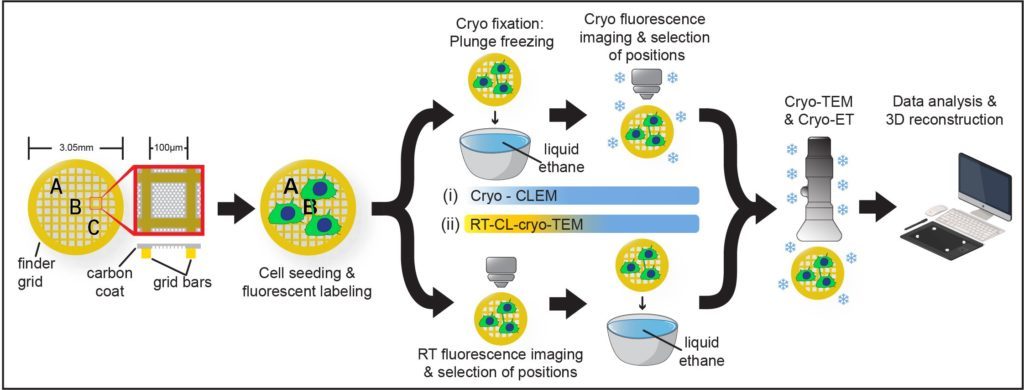
Furthermore, by setting up a workflow in cryo-CLEM we have demonstrated that they constitute novel open-ended structures supporting the passage of organelles identified by tomography and live imaging (Sartori*, Cordero*, Pepe et al., Nat comm 2019).
In this project we will undertake a comprehensive approach by employing interdisciplinary tools (mathematics, machine learning, biophysics, state-of-the-art imaging techniques, from high-throughput electron to light-sheet microscopy, biochemistry and cell/molecular biology) in several models (microfluidic cultures, mouse brain, human organoids and zebra-fish) to: i) uncover the biophysical/mechanical characteristics of TNTs; ii) identify specific pathways and TNT markers; iii) demonstrate their existence in vivo; and iv) analyze their role in the progression of cancer and NDs.
Specifically, we are using micropatterning and microfluidics to investigate: 1) the mechanisms of formation and biophysical characteristics of TNTs; 2) the role of actin modulators in TNT formation, dynamics and mechanical properties; 3) the mechanisms of fusion in TNTs.
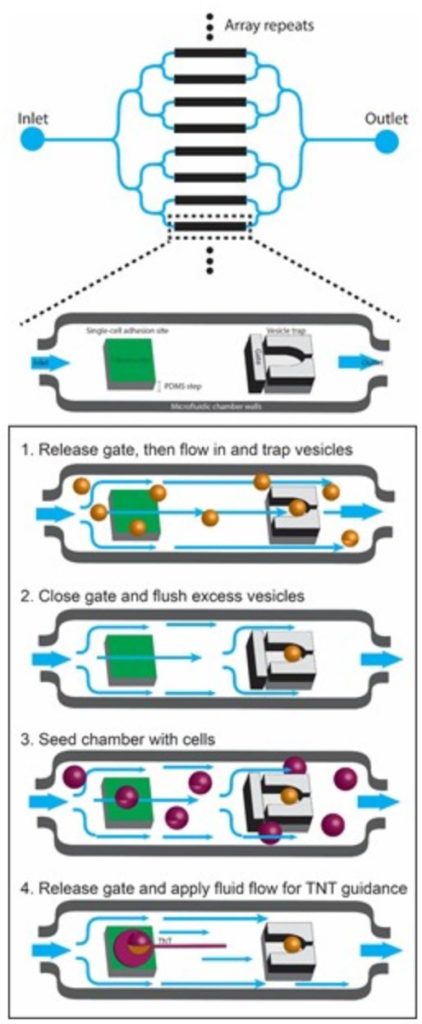
And we will use machine learning for high-content quantification of TNTs in vitro. Finally, we are investigating the formation and function of TNTs in vivo during development in different models: zebra fish embryo, mouse brain and human organoids, using a combination of high-resolution microscopy techniques including live imaging and lattice light-sheet microscopy, cryo-CLEM tomography and serial sectioning scanning electron microscopy (ssSEM) and cell segmentation, combined with different analytical imaging software q-for quantitative image analysis
Overall this project will decipher the biophysical characteristics of TNTs and the mechanisms governing TNT formation and fusion. It will identify TNT-specific pathways and biomarkers and will set up high-throughput quantifications currently lacking and absolutely necessary for their quantitative in vivo study. It will finally provide the demonstration of the existence and function of TNTs in vivo, pioneering further studies in different physiological conditions and models of diseases.