Présentation
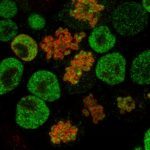
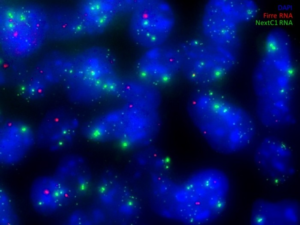
The study of ES cells has not escaped the recent explosion of regulatory lncRNA biology: from the thousands of lncRNAs that have been identified across the mouse genome a large subset is expressed in ES cells. Our goal is to identify “pluripotency lncRNAs” that would impact on ES cell biology as strongly as well established pluripotency transcription factors. We will focus on Nanog-mediated regulation as a tool to discover relevant lncRNAs. Indeed, among the three main pluripotency factors (Oct4, Sox2 and Nanog), only Nanog can be manipulated without dramatically affecting the ES cell transcriptome. Yet, within the very limited number of genes strictly dependent upon Nanog, highly relevant transcription factors such as Esrrb, Klf4, Rex1 and Prdm14 are found. We therefore postulate that within the potentially small number of Nanog-dependent lncRNAs, molecules as important as the aforementioned transcription factors should be easily identified. We are using RNA-Seq approaches to identify lncRNAs (LASR for Long-non-coding Associated to Self-Renewal) that respond to Nanog levels and are associated to the ground state of pluripotency. We are testing their functions using a combination of CRISPR-activation and -deletion studies.